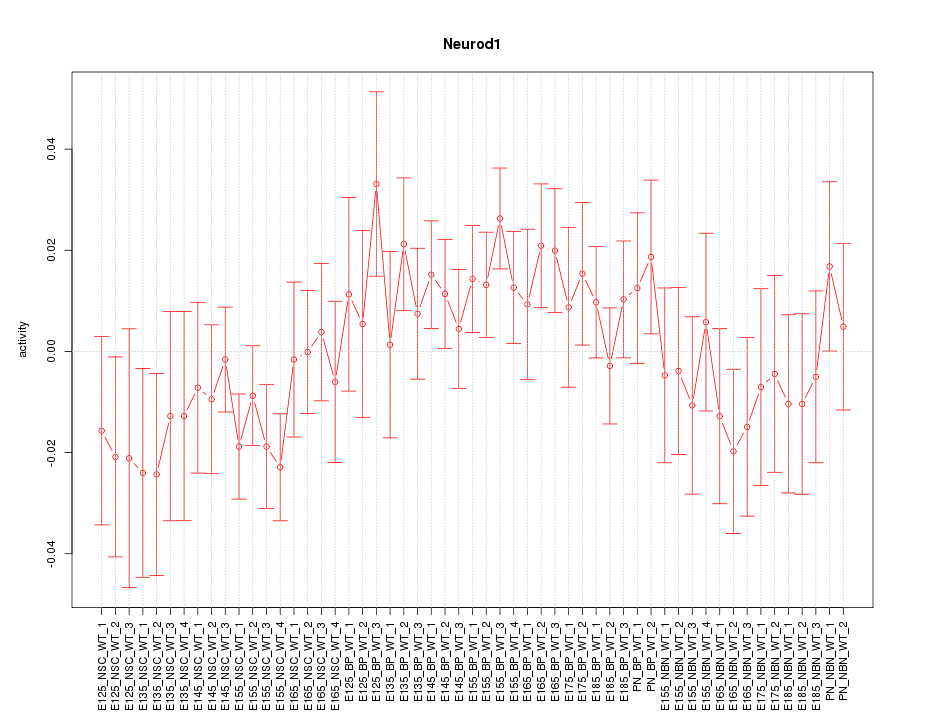
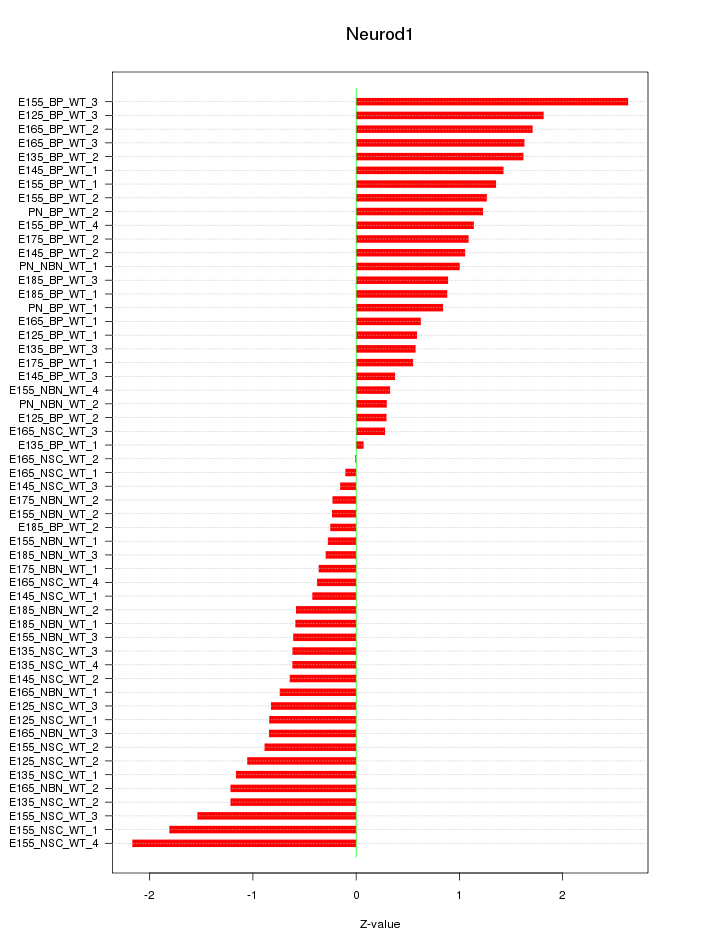
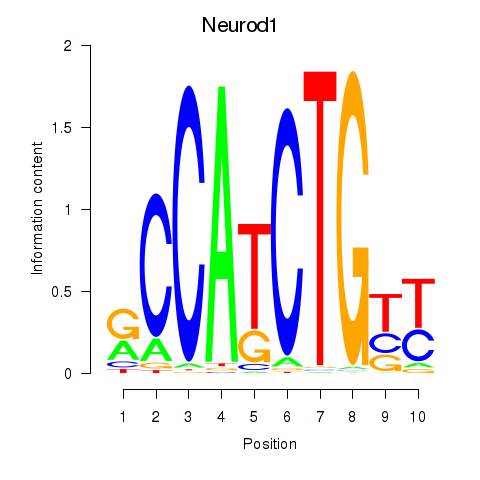
Motif ID: Neurod1
Z-value: 1.013
Transcription factors associated with Neurod1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Neurod1 | ENSMUSG00000034701.9 | Neurod1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Neurod1 | mm10_v2_chr2_-_79456750_79456761 | 0.62 | 4.5e-07 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.4 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.6 | 3.4 | GO:0097473 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.5 | 2.6 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.5 | 1.9 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.5 | 4.6 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.5 | 5.0 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.4 | 1.3 | GO:0039533 | regulation of MDA-5 signaling pathway(GO:0039533) |
| 0.4 | 2.4 | GO:0003383 | apical constriction(GO:0003383) |
| 0.4 | 3.6 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.4 | 2.0 | GO:0046864 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) alveolar primary septum development(GO:0061143) |
| 0.3 | 1.0 | GO:0090425 | hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.3 | 1.6 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.3 | 0.9 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 0.3 | 1.5 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.3 | 1.1 | GO:1904451 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.3 | 2.4 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.3 | 1.1 | GO:1902340 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.2 | 2.9 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.2 | 0.7 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.2 | 1.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 | 1.7 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.2 | 1.5 | GO:0044838 | cell quiescence(GO:0044838) |
| 0.2 | 1.6 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.2 | 1.2 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.2 | 2.9 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.2 | 1.8 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.2 | 1.8 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.2 | 2.7 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.2 | 1.0 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.2 | 0.5 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.2 | 0.5 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.2 | 2.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.2 | 6.8 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 1.7 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 | 2.2 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.2 | 1.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.2 | 0.6 | GO:0099624 | atrial cardiac muscle cell membrane repolarization(GO:0099624) |
| 0.2 | 0.6 | GO:0060032 | notochord regression(GO:0060032) |
| 0.1 | 0.7 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.1 | 0.9 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.1 | 1.7 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.1 | 1.0 | GO:0035826 | rubidium ion transport(GO:0035826) cellular hypotonic response(GO:0071476) |
| 0.1 | 1.5 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.7 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.7 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 2.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 0.9 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.1 | 1.0 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 1.1 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.1 | 0.6 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.1 | 1.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 1.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.7 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.1 | 0.5 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.6 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 2.7 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 1.0 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) prepulse inhibition(GO:0060134) |
| 0.1 | 0.3 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.1 | 0.5 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 | 0.3 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) excitatory synapse assembly(GO:1904861) |
| 0.1 | 1.5 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 0.5 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.1 | 0.4 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 1.4 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 0.7 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 0.7 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.1 | 0.3 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.1 | 1.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 0.6 | GO:0043586 | tongue development(GO:0043586) |
| 0.1 | 1.7 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.1 | 0.5 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.9 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 1.4 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.1 | 0.3 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 0.4 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.1 | 0.8 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 1.4 | GO:0007617 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.1 | 0.3 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 | 0.9 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.1 | 1.0 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.1 | 0.7 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 1.6 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.5 | GO:0090003 | regulation of Golgi to plasma membrane protein transport(GO:0042996) regulation of establishment of protein localization to plasma membrane(GO:0090003) |
| 0.0 | 1.2 | GO:0001523 | retinoid metabolic process(GO:0001523) |
| 0.0 | 0.2 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.3 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.1 | GO:0006778 | porphyrin-containing compound metabolic process(GO:0006778) porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 0.5 | GO:1901898 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 1.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.3 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.2 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.5 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 3.4 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.7 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.4 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 1.1 | GO:0035136 | forelimb morphogenesis(GO:0035136) |
| 0.0 | 1.6 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 4.2 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.0 | 1.5 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.4 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 0.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.3 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.6 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.9 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.2 | GO:0048149 | adult feeding behavior(GO:0008343) behavioral response to ethanol(GO:0048149) |
| 0.0 | 1.0 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.2 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 0.7 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 2.9 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.0 | 0.4 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 1.0 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.2 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.2 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) |
| 0.0 | 0.4 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.2 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.3 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.7 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.7 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.4 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 3.3 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 1.6 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 0.1 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 0.5 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 0.4 | GO:0086001 | cardiac muscle cell action potential(GO:0086001) |
| 0.0 | 0.2 | GO:0002192 | IRES-dependent translational initiation(GO:0002192) IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 2.2 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.7 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 0.2 | GO:1903861 | regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.2 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.5 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.0 | 0.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.1 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.1 | GO:0036159 | outer dynein arm assembly(GO:0036158) inner dynein arm assembly(GO:0036159) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0090537 | CERF complex(GO:0090537) |
| 0.3 | 1.1 | GO:0097574 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) |
| 0.3 | 1.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.2 | 0.9 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.2 | 3.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 3.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.2 | 1.4 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.2 | 5.0 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.2 | 1.6 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 6.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 1.7 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.1 | 1.0 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 1.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.5 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 0.4 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.1 | 0.9 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 0.3 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 0.9 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 2.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 2.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 5.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 0.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.9 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 2.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 4.5 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.1 | 0.4 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 2.9 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 1.1 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.6 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.4 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.8 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 1.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 1.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.6 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 1.7 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 2.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 2.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.3 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 2.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 3.1 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 1.0 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 4.2 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.2 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 1.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.7 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 2.6 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.7 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.6 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 1.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 1.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.6 | 1.7 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.5 | 1.5 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.4 | 2.6 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.4 | 1.8 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.4 | 2.9 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.3 | 6.4 | GO:0001848 | complement binding(GO:0001848) |
| 0.3 | 1.5 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.3 | 1.5 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.3 | 2.2 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.3 | 3.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 1.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 0.7 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.2 | 5.3 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 1.3 | GO:0089720 | caspase binding(GO:0089720) |
| 0.2 | 0.6 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.2 | 1.7 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 1.7 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.2 | 1.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 2.4 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.2 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.2 | 6.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.2 | 0.9 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 0.9 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 1.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 1.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 2.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 1.0 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 1.0 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 1.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 1.6 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.1 | 0.7 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 0.6 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.9 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.5 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 1.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.6 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.1 | 2.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 2.9 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 0.5 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 0.5 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 4.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 2.9 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 1.4 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.1 | 2.1 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 4.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.4 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.3 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.1 | 2.0 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.1 | 0.4 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.4 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 1.0 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 1.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 1.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.7 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 2.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 6.9 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.2 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.7 | GO:0071949 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) FAD binding(GO:0071949) |
| 0.0 | 0.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.6 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 1.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.5 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 1.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.7 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 1.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.4 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.2 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.3 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.9 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.1 | PID_TCR_JNK_PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 6.6 | ST_GA13_PATHWAY | G alpha 13 Pathway |
| 0.1 | 1.7 | PID_DNA_PK_PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 0.2 | PID_RANBP2_PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 2.0 | NABA_BASEMENT_MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.1 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.0 | 1.0 | PID_IL2_STAT5_PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.8 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 4.1 | PID_HDAC_CLASSI_PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 2.7 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 2.0 | PID_IL2_1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.0 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | PID_TCR_RAS_PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.9 | PID_CERAMIDE_PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.7 | PID_IL6_7_PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.4 | SIG_INSULIN_RECEPTOR_PATHWAY_IN_CARDIAC_MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.6 | PID_RET_PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.6 | PID_ERBB4_PATHWAY | ErbB4 signaling events |
| 0.0 | 2.2 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.3 | PID_NOTCH_PATHWAY | Notch signaling pathway |
| 0.0 | 1.5 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.6 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 6.4 | REACTOME_INTRINSIC_PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 3.4 | REACTOME_NEGATIVE_REGULATION_OF_THE_PI3K_AKT_NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.9 | REACTOME_REGULATION_OF_GENE_EXPRESSION_IN_BETA_CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 2.4 | REACTOME_PRE_NOTCH_PROCESSING_IN_GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 2.6 | REACTOME_TRAFFICKING_OF_GLUR2_CONTAINING_AMPA_RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 3.7 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.8 | REACTOME_PLATELET_SENSITIZATION_BY_LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 1.0 | REACTOME_CYTOCHROME_P450_ARRANGED_BY_SUBSTRATE_TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 3.7 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELLULAR_PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 1.2 | REACTOME_UNBLOCKING_OF_NMDA_RECEPTOR_GLUTAMATE_BINDING_AND_ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 4.6 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 2.4 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.4 | REACTOME_TGF_BETA_RECEPTOR_SIGNALING_IN_EMT_EPITHELIAL_TO_MESENCHYMAL_TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.5 | REACTOME_AMINO_ACID_SYNTHESIS_AND_INTERCONVERSION_TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.7 | REACTOME_SYNTHESIS_OF_PE | Genes involved in Synthesis of PE |
| 0.1 | 0.5 | REACTOME_PLATELET_ADHESION_TO_EXPOSED_COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.6 | REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 1.5 | REACTOME_GROWTH_HORMONE_RECEPTOR_SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.5 | REACTOME_ROLE_OF_DCC_IN_REGULATING_APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 1.1 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.9 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.1 | REACTOME_RAP1_SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME_RECRUITMENT_OF_NUMA_TO_MITOTIC_CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.5 | REACTOME_HORMONE_SENSITIVE_LIPASE_HSL_MEDIATED_TRIACYLGLYCEROL_HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME_GAP_JUNCTION_ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.2 | REACTOME_AKT_PHOSPHORYLATES_TARGETS_IN_THE_CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 1.9 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.3 | REACTOME_APOPTOSIS_INDUCED_DNA_FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.7 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.7 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.0 | 0.5 | REACTOME_REGULATION_OF_SIGNALING_BY_CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 1.8 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.7 | REACTOME_SIGNALING_BY_ROBO_RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.0 | REACTOME_POTASSIUM_CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 1.2 | REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.5 | REACTOME_SMOOTH_MUSCLE_CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME_KERATAN_SULFATE_BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |