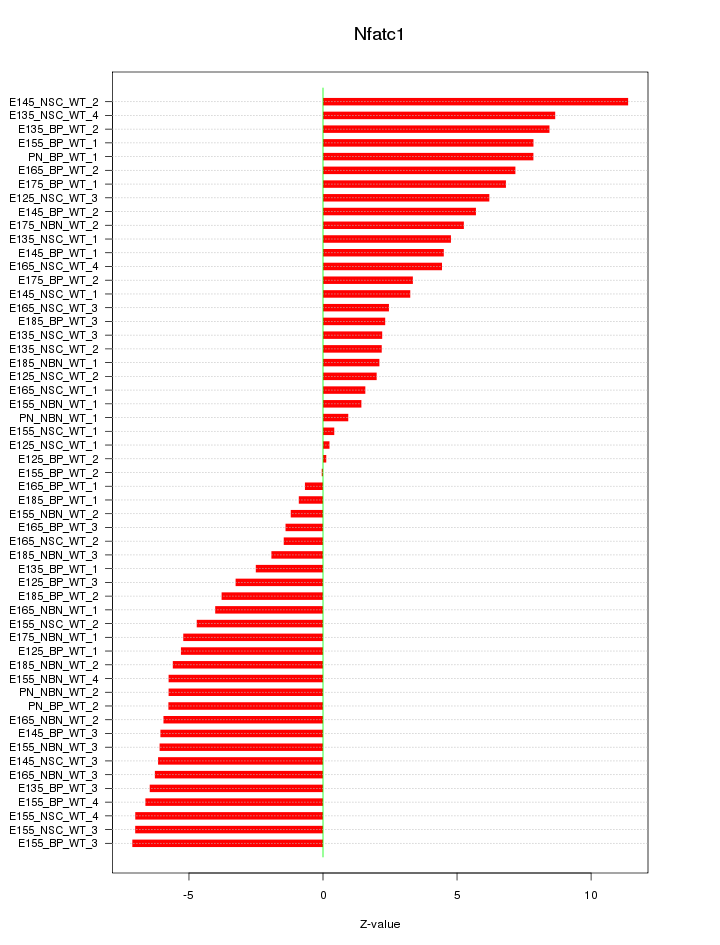
Motif ID: Nfatc1
Z-value: 5.051
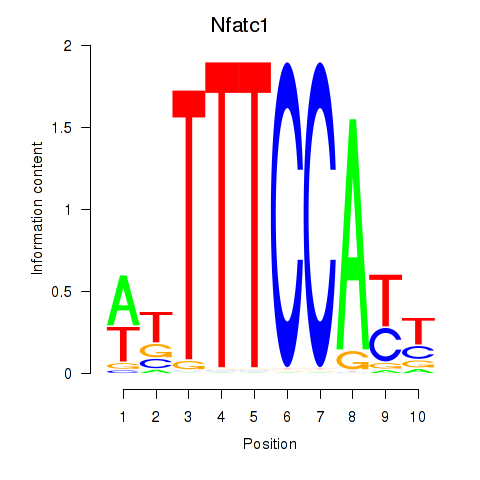
Transcription factors associated with Nfatc1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nfatc1 | ENSMUSG00000033016.9 | Nfatc1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfatc1 | mm10_v2_chr18_-_80708115_80708180 | 0.12 | 3.7e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.4 | 25.1 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 5.8 | 17.3 | GO:0060745 | mammary gland branching involved in pregnancy(GO:0060745) |
| 5.6 | 16.8 | GO:0035622 | intrahepatic bile duct development(GO:0035622) renal vesicle induction(GO:0072034) |
| 4.8 | 14.5 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 4.2 | 34.0 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 3.6 | 10.8 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 3.4 | 17.1 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 3.4 | 23.5 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 3.2 | 9.7 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 2.5 | 12.6 | GO:1901526 | negative regulation of mitochondrial membrane permeability(GO:0035795) positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 2.1 | 6.4 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 1.6 | 4.7 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 1.6 | 12.5 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 1.5 | 6.2 | GO:1903416 | negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) regulation of calcidiol 1-monooxygenase activity(GO:0060558) response to glycoside(GO:1903416) |
| 1.5 | 10.5 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 1.4 | 5.5 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 1.3 | 5.3 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 1.3 | 11.4 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 1.2 | 15.8 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 1.1 | 6.7 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 1.1 | 6.5 | GO:0002317 | plasma cell differentiation(GO:0002317) positive regulation of viral entry into host cell(GO:0046598) |
| 1.1 | 2.1 | GO:0060126 | somatotropin secreting cell differentiation(GO:0060126) |
| 1.0 | 11.0 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.9 | 8.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.9 | 5.5 | GO:1904587 | response to glycoprotein(GO:1904587) |
| 0.9 | 10.6 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.8 | 3.3 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.8 | 4.8 | GO:0035989 | tendon development(GO:0035989) |
| 0.8 | 4.7 | GO:0098734 | protein depalmitoylation(GO:0002084) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) negative regulation of establishment of protein localization to plasma membrane(GO:0090005) macromolecule depalmitoylation(GO:0098734) |
| 0.8 | 3.1 | GO:2000794 | regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.8 | 2.3 | GO:0090135 | actin filament branching(GO:0090135) negative regulation of phospholipase C activity(GO:1900275) |
| 0.8 | 8.5 | GO:0072189 | ureter development(GO:0072189) |
| 0.7 | 2.2 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.7 | 8.0 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.7 | 7.3 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.6 | 1.9 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) negative regulation of Fas signaling pathway(GO:1902045) |
| 0.6 | 1.9 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.6 | 9.9 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.6 | 1.9 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.6 | 1.9 | GO:2000373 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.6 | 2.3 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.5 | 2.5 | GO:0060353 | regulation of cell adhesion molecule production(GO:0060353) renal water absorption(GO:0070295) |
| 0.5 | 1.9 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.5 | 2.8 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.4 | 2.7 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.4 | 1.8 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.4 | 2.1 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.4 | 3.3 | GO:0097152 | mesenchymal cell apoptotic process(GO:0097152) |
| 0.4 | 1.6 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.4 | 6.7 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.4 | 1.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.4 | 1.9 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.4 | 17.5 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.4 | 2.2 | GO:0061009 | common bile duct development(GO:0061009) |
| 0.4 | 7.8 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.3 | 1.3 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.3 | 4.7 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.3 | 1.4 | GO:1900169 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.3 | 3.1 | GO:0043586 | tongue development(GO:0043586) |
| 0.3 | 1.6 | GO:0032439 | endosome localization(GO:0032439) |
| 0.3 | 0.8 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.3 | 3.6 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.2 | 5.0 | GO:1903077 | negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.2 | 6.9 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.2 | 1.2 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.2 | 5.0 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.2 | 8.7 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.2 | 6.5 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.2 | 1.5 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.2 | 1.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.2 | 0.9 | GO:0019659 | fermentation(GO:0006113) lactate biosynthetic process from pyruvate(GO:0019244) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.2 | 4.8 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.2 | 0.8 | GO:0021856 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) cranial ganglion development(GO:0061550) trigeminal ganglion development(GO:0061551) facioacoustic ganglion development(GO:1903375) |
| 0.2 | 2.7 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.2 | 2.2 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.2 | 2.9 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.2 | 2.3 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 8.6 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 1.4 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 1.5 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 5.1 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.1 | 0.3 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.1 | 5.0 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.8 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 2.6 | GO:0060441 | epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.1 | 2.1 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.1 | 6.1 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.1 | 1.0 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 1.4 | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.1 | 3.0 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 3.2 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.1 | 1.9 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 3.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 3.5 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 10.8 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.1 | 0.5 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 0.4 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 3.7 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 2.0 | GO:0043153 | photoperiodism(GO:0009648) entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 0.3 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 2.0 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 1124.6 | GO:0008150 | biological_process(GO:0008150) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 14.5 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 3.2 | 9.7 | GO:0071914 | prominosome(GO:0071914) |
| 1.8 | 12.4 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 1.3 | 17.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.9 | 6.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.7 | 2.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.6 | 8.4 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.5 | 8.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.4 | 6.7 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.4 | 1.3 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.4 | 4.8 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.4 | 1.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.3 | 4.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.3 | 3.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.3 | 2.8 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.3 | 1.9 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.3 | 4.9 | GO:0036038 | MKS complex(GO:0036038) |
| 0.3 | 1.6 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 1.5 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.2 | 3.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 2.7 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.2 | 2.9 | GO:0042581 | specific granule(GO:0042581) |
| 0.2 | 11.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 7.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 9.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 2.3 | GO:0051286 | cell tip(GO:0051286) |
| 0.1 | 2.8 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 24.8 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.1 | 3.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 25.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 9.7 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.1 | 4.2 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.5 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 5.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 1453.1 | GO:0005575 | cellular_component(GO:0005575) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.4 | 33.6 | GO:0008142 | oxysterol binding(GO:0008142) |
| 5.8 | 17.3 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 4.2 | 12.5 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 3.7 | 14.9 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 3.6 | 14.5 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 2.5 | 12.6 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 2.2 | 6.7 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 1.7 | 18.3 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 1.3 | 6.5 | GO:0005534 | galactose binding(GO:0005534) |
| 1.2 | 4.7 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 1.1 | 6.4 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 1.0 | 9.0 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.9 | 5.4 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.9 | 18.8 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.8 | 5.3 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.7 | 23.0 | GO:0042805 | actinin binding(GO:0042805) |
| 0.6 | 1.9 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.6 | 7.8 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.6 | 5.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.5 | 4.7 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.5 | 1.9 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.5 | 5.5 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.4 | 1.3 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.4 | 1.6 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.4 | 5.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.4 | 1.9 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.4 | 7.3 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.4 | 3.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.4 | 2.9 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.4 | 1.4 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.4 | 2.5 | GO:0015288 | porin activity(GO:0015288) |
| 0.3 | 22.3 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.3 | 1.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.3 | 1.3 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.3 | 8.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 0.9 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.2 | 1.5 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 2.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.2 | 1.9 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.2 | 5.5 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.2 | 1.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.2 | 5.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 2.5 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.2 | 0.9 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.2 | 3.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 1.2 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.2 | 3.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 0.5 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 4.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 4.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 2.1 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 5.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 23.4 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 2.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 1.9 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 2.3 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 0.8 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 1.9 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 1.5 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.1 | 1318.0 | GO:0003674 | molecular_function(GO:0003674) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 17.1 | PID_INTEGRIN2_PATHWAY | Beta2 integrin cell surface interactions |
| 0.9 | 49.3 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.8 | 5.3 | ST_PAC1_RECEPTOR_PATHWAY | PAC1 Receptor Pathway |
| 0.6 | 17.9 | PID_UPA_UPAR_PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.5 | 30.4 | PID_IL12_2PATHWAY | IL12-mediated signaling events |
| 0.4 | 16.4 | PID_INSULIN_GLUCOSE_PATHWAY | Insulin-mediated glucose transport |
| 0.3 | 22.3 | PID_P73PATHWAY | p73 transcription factor network |
| 0.2 | 10.8 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.2 | 15.4 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 1.9 | PID_INTEGRIN_A9B1_PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.2 | 1.9 | PID_THROMBIN_PAR4_PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 6.9 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 6.0 | PID_FCER1_PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 5.1 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 5.3 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.1 | 4.7 | PID_ANGIOPOIETIN_RECEPTOR_PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 1.9 | PID_GLYPICAN_1PATHWAY | Glypican 1 network |
| 0.1 | 13.2 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.3 | PID_IL6_7_PATHWAY | IL6-mediated signaling events |
| 0.1 | 3.4 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
| 0.1 | 2.1 | PID_RHOA_PATHWAY | RhoA signaling pathway |
| 0.1 | 2.8 | PID_HEDGEHOG_GLI_PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.3 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 2.6 | ST_FAS_SIGNALING_PATHWAY | Fas Signaling Pathway |
| 0.0 | 3.5 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 2.0 | PID_CMYB_PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.4 | PID_RHOA_REG_PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.3 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.3 | PID_ATF2_PATHWAY | ATF-2 transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | REACTOME_HORMONE_LIGAND_BINDING_RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 1.5 | 14.5 | REACTOME_G2_M_DNA_DAMAGE_CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.9 | 23.5 | REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.8 | 57.6 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.5 | 3.1 | REACTOME_RNA_POL_III_TRANSCRIPTION_TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.5 | 5.5 | REACTOME_PROLACTIN_RECEPTOR_SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.3 | 10.5 | REACTOME_RNA_POL_III_TRANSCRIPTION_INITIATION_FROM_TYPE_3_PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.3 | 8.4 | REACTOME_DCC_MEDIATED_ATTRACTIVE_SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.3 | 2.5 | REACTOME_PASSIVE_TRANSPORT_BY_AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 12.6 | REACTOME_GLUCOSE_TRANSPORT | Genes involved in Glucose transport |
| 0.3 | 12.5 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.2 | 6.3 | REACTOME_TIGHT_JUNCTION_INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 4.5 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 5.2 | REACTOME_ENOS_ACTIVATION_AND_REGULATION | Genes involved in eNOS activation and regulation |
| 0.2 | 6.7 | REACTOME_INHIBITION_OF_VOLTAGE_GATED_CA2_CHANNELS_VIA_GBETA_GAMMA_SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 2.1 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 0.1 | 3.2 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.9 | REACTOME_METABOLISM_OF_POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.7 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 6.5 | REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 0.8 | REACTOME_SEMA3A_PLEXIN_REPULSION_SIGNALING_BY_INHIBITING_INTEGRIN_ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 1.9 | REACTOME_SEMA4D_INDUCED_CELL_MIGRATION_AND_GROWTH_CONE_COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.6 | REACTOME_AQUAPORIN_MEDIATED_TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.0 | 5.6 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 10.7 | REACTOME_ANTIGEN_PROCESSING_UBIQUITINATION_PROTEASOME_DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.6 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.9 | REACTOME_PYRUVATE_METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.3 | REACTOME_GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 1.8 | REACTOME_TRANSCRIPTIONAL_REGULATION_OF_WHITE_ADIPOCYTE_DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.7 | REACTOME_PACKAGING_OF_TELOMERE_ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.3 | REACTOME_METABOLISM_OF_STEROID_HORMONES_AND_VITAMINS_A_AND_D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 2.0 | REACTOME_MRNA_SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.5 | REACTOME_RECYCLING_PATHWAY_OF_L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.6 | REACTOME_LATE_PHASE_OF_HIV_LIFE_CYCLE | Genes involved in Late Phase of HIV Life Cycle |