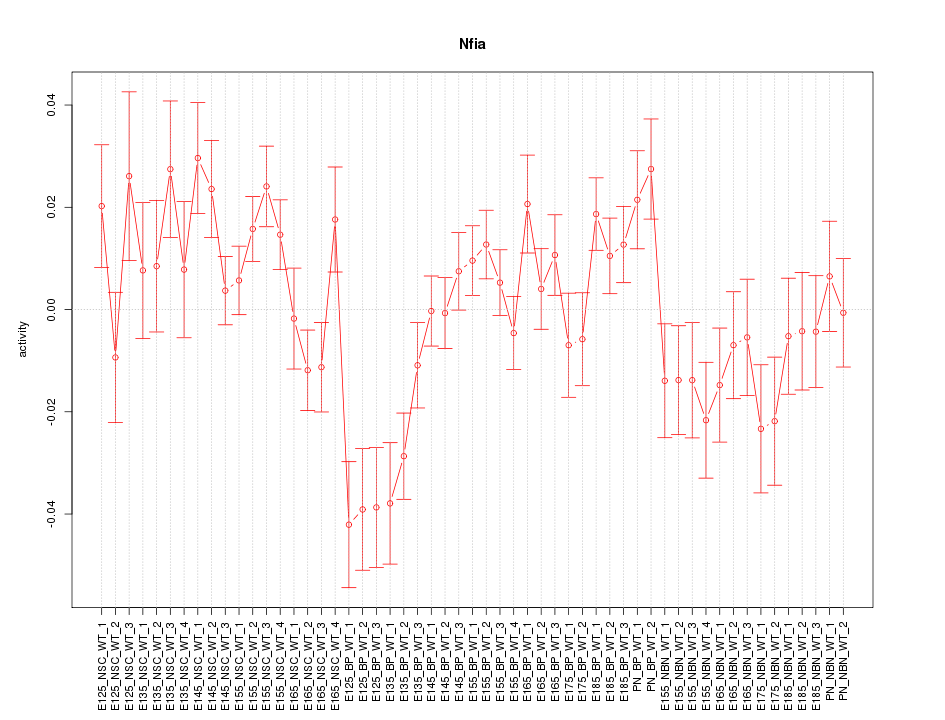
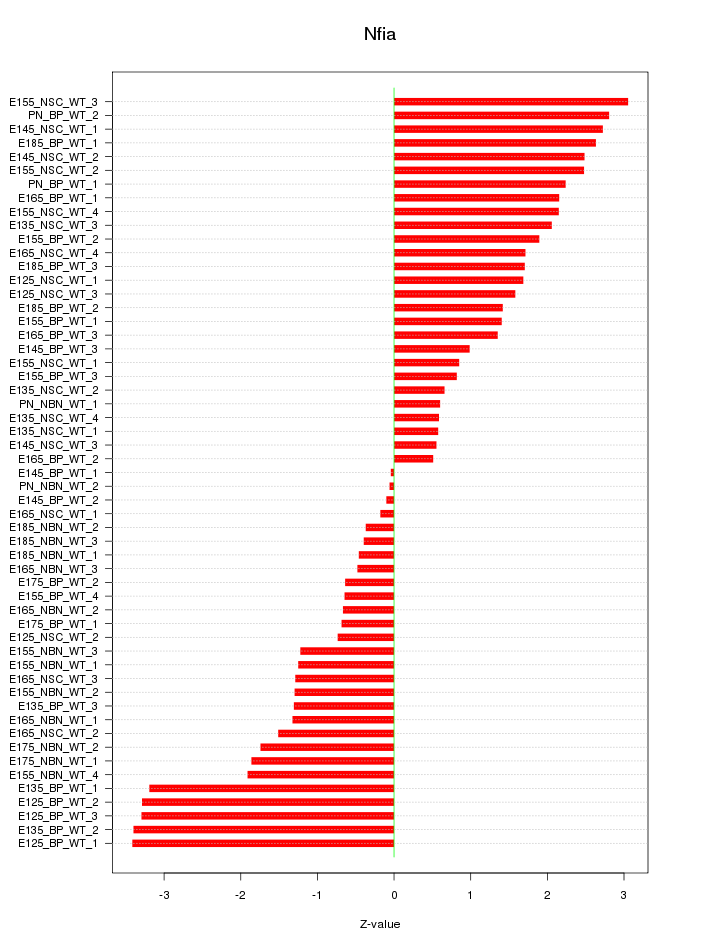
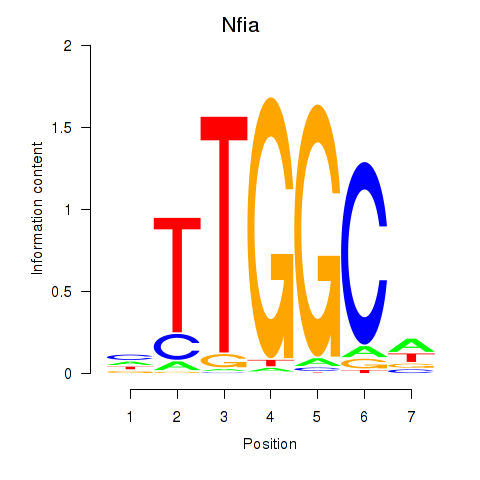
Motif ID: Nfia
Z-value: 1.747
Transcription factors associated with Nfia:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nfia | ENSMUSG00000028565.12 | Nfia |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfia | mm10_v2_chr4_+_97777780_97777834 | 0.27 | 4.8e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 15.7 | GO:1902460 | transforming growth factor beta activation(GO:0036363) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 3.9 | 19.3 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 3.4 | 10.1 | GO:0045168 | cell-cell signaling involved in cell fate commitment(GO:0045168) |
| 3.0 | 6.0 | GO:0072197 | ureter morphogenesis(GO:0072197) |
| 2.9 | 8.8 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 2.7 | 2.7 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 2.7 | 5.3 | GO:0072138 | mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) |
| 2.3 | 13.8 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 2.2 | 15.6 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 2.2 | 8.7 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 2.2 | 6.5 | GO:0097350 | neutrophil clearance(GO:0097350) |
| 2.1 | 6.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 2.1 | 6.2 | GO:0060737 | prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) |
| 1.9 | 19.0 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 1.9 | 7.6 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 1.8 | 7.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) cellular response to lipid hydroperoxide(GO:0071449) |
| 1.7 | 6.9 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 1.6 | 3.3 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 1.6 | 7.9 | GO:0021905 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 1.6 | 7.9 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 1.5 | 4.6 | GO:0090425 | hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 1.5 | 4.6 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) |
| 1.5 | 3.1 | GO:1905065 | positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 1.5 | 6.0 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 1.5 | 7.5 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 1.4 | 8.7 | GO:0010624 | regulation of Schwann cell proliferation(GO:0010624) |
| 1.4 | 5.5 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) bud outgrowth involved in lung branching(GO:0060447) |
| 1.4 | 6.9 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 1.3 | 1.3 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 1.2 | 3.6 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 1.2 | 4.6 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 1.1 | 3.4 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 1.1 | 12.5 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 1.1 | 3.3 | GO:0009946 | proximal/distal axis specification(GO:0009946) neuroblast migration(GO:0097402) |
| 1.1 | 6.6 | GO:0032196 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) transposition(GO:0032196) |
| 1.1 | 8.6 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 1.1 | 9.7 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 1.0 | 1.0 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 1.0 | 10.0 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.9 | 2.8 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.9 | 4.7 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.9 | 1.8 | GO:0034759 | regulation of iron ion transport(GO:0034756) regulation of iron ion transmembrane transport(GO:0034759) |
| 0.9 | 2.7 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.9 | 2.6 | GO:1902524 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.9 | 3.4 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.9 | 3.4 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.8 | 0.8 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.8 | 0.8 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.8 | 2.5 | GO:0032650 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) cardiac cell fate determination(GO:0060913) |
| 0.8 | 4.0 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.8 | 3.2 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.8 | 0.8 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.8 | 0.8 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.8 | 2.3 | GO:0002030 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) |
| 0.8 | 3.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.8 | 7.8 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.8 | 3.9 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.8 | 3.8 | GO:0052428 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.8 | 0.8 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.8 | 3.0 | GO:0002484 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.7 | 2.2 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.7 | 7.9 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.7 | 2.9 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.7 | 3.4 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.7 | 1.4 | GO:1903181 | regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 0.7 | 8.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.7 | 2.0 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.7 | 1.3 | GO:0036065 | fucosylation(GO:0036065) |
| 0.7 | 3.3 | GO:0042891 | antibiotic transport(GO:0042891) dipeptide transport(GO:0042938) |
| 0.7 | 2.0 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.7 | 2.6 | GO:1902724 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.6 | 2.6 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.6 | 1.9 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.6 | 1.9 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.6 | 3.1 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.6 | 1.8 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.6 | 6.5 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.6 | 2.4 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.6 | 11.2 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.6 | 2.4 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.6 | 1.8 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.6 | 1.7 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.5 | 1.6 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.5 | 3.2 | GO:0097473 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.5 | 0.5 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.5 | 5.3 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.5 | 1.1 | GO:0070343 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 0.5 | 4.1 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.5 | 0.5 | GO:0097278 | complement-dependent cytotoxicity(GO:0097278) |
| 0.5 | 1.5 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.5 | 8.6 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.5 | 2.0 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.5 | 1.5 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.5 | 1.4 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.5 | 1.0 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.5 | 1.9 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.5 | 2.8 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.5 | 3.7 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.5 | 2.7 | GO:1902998 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.5 | 2.7 | GO:0032346 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.4 | 1.3 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) DNA replication preinitiation complex assembly(GO:0071163) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.4 | 1.3 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.4 | 1.7 | GO:0019323 | D-ribose metabolic process(GO:0006014) pentose catabolic process(GO:0019323) |
| 0.4 | 1.3 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.4 | 1.2 | GO:0009153 | purine deoxyribonucleotide biosynthetic process(GO:0009153) |
| 0.4 | 2.4 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.4 | 3.9 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.4 | 1.2 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.4 | 0.8 | GO:2000468 | regulation of peroxidase activity(GO:2000468) |
| 0.4 | 1.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.3 | 1.0 | GO:0048377 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) positive regulation of cardiac ventricle development(GO:1904414) |
| 0.3 | 3.8 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.3 | 2.4 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.3 | 5.4 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.3 | 1.3 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.3 | 0.7 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.3 | 2.3 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.3 | 1.0 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.3 | 1.0 | GO:0061428 | embryonic heart tube left/right pattern formation(GO:0060971) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.3 | 1.0 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.3 | 2.3 | GO:0046549 | phenol-containing compound catabolic process(GO:0019336) retinal cone cell development(GO:0046549) |
| 0.3 | 3.2 | GO:0051231 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) spindle midzone assembly(GO:0051255) mitotic spindle midzone assembly(GO:0051256) |
| 0.3 | 3.5 | GO:0072112 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.3 | 1.6 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.3 | 0.3 | GO:1902512 | positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.3 | 4.3 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.3 | 0.9 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.3 | 0.6 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.3 | 1.5 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.3 | 0.9 | GO:0060596 | regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003256) mammary placode formation(GO:0060596) |
| 0.3 | 0.6 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.3 | 1.2 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.3 | 3.7 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.3 | 0.9 | GO:0038095 | positive regulation of mast cell cytokine production(GO:0032765) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.3 | 2.3 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.3 | 1.4 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.3 | 1.4 | GO:1904721 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.3 | 0.8 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.3 | 0.8 | GO:0021972 | corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.3 | 0.5 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.3 | 0.5 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.3 | 0.3 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.3 | 0.3 | GO:0002265 | astrocyte activation involved in immune response(GO:0002265) |
| 0.3 | 3.4 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.3 | 0.5 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.3 | 0.5 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.3 | 1.0 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.3 | 6.4 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.3 | 1.3 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.3 | 1.8 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.2 | 0.5 | GO:0010957 | negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.2 | 1.0 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.2 | 2.4 | GO:0002118 | aggressive behavior(GO:0002118) |
| 0.2 | 1.0 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) dibasic protein processing(GO:0090472) |
| 0.2 | 1.2 | GO:1903624 | regulation of apoptotic DNA fragmentation(GO:1902510) regulation of DNA catabolic process(GO:1903624) |
| 0.2 | 1.7 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.2 | 0.9 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.2 | 6.5 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.2 | 0.7 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.2 | 0.9 | GO:0015744 | succinate transport(GO:0015744) |
| 0.2 | 1.8 | GO:0042095 | interferon-gamma biosynthetic process(GO:0042095) |
| 0.2 | 1.6 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.2 | 1.1 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.2 | 0.7 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.2 | 0.9 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.2 | 7.1 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.2 | 0.4 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.2 | 1.5 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.2 | 6.9 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 1.5 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.2 | 0.6 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.2 | 1.3 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.2 | 7.5 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.2 | 1.7 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.2 | 0.6 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.2 | 1.3 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.2 | 1.1 | GO:1905169 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.2 | 0.8 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.2 | 5.1 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.2 | 2.6 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.2 | 1.6 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.2 | 2.0 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.2 | 0.8 | GO:0060266 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.2 | 0.4 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.2 | 1.3 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.2 | 4.0 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.2 | 4.1 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.2 | 1.1 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.2 | 1.2 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.2 | 1.1 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.2 | 0.5 | GO:0019230 | proprioception(GO:0019230) sensory neuron axon guidance(GO:0097374) |
| 0.2 | 1.0 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.2 | 1.7 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 1.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 3.2 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.2 | 0.7 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.2 | 2.6 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.2 | 0.7 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.2 | 7.5 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.2 | 1.6 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.2 | 3.9 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.2 | 0.6 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.2 | 3.3 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.2 | 3.1 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.2 | 4.8 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.2 | 0.9 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.2 | 1.8 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.2 | 0.9 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.2 | 0.9 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.2 | 0.3 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.2 | 1.7 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.2 | 0.5 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.1 | 1.0 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.1 | 1.2 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.1 | 0.1 | GO:1903984 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.1 | 2.9 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.1 | 2.5 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 0.4 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 0.1 | 0.6 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.1 | 0.6 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.1 | 0.6 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 0.6 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 1.6 | GO:0014883 | transition between fast and slow fiber(GO:0014883) lateral mesoderm development(GO:0048368) |
| 0.1 | 0.4 | GO:0003340 | negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003340) |
| 0.1 | 0.4 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.1 | 1.7 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 3.5 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 2.0 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.1 | 1.6 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 2.7 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.1 | 1.7 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 2.7 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.1 | 0.4 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.1 | 1.7 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 0.2 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 | 1.4 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.1 | 0.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.5 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.1 | 0.5 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 1.7 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 1.2 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.8 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.1 | 0.5 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.1 | 0.7 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 1.4 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 0.6 | GO:0060347 | heart trabecula formation(GO:0060347) |
| 0.1 | 1.1 | GO:0070303 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of JNK cascade(GO:0046329) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.1 | 0.3 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.3 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 0.3 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.1 | 0.8 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.7 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 1.4 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 0.7 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.6 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.4 | GO:1903286 | regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 2.6 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.1 | 0.7 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 | 0.4 | GO:0055009 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.1 | 0.5 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.1 | 0.2 | GO:1904245 | regulation of polynucleotide adenylyltransferase activity(GO:1904245) |
| 0.1 | 1.3 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.1 | 0.6 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 0.4 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.1 | 0.4 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.5 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.4 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.1 | 0.6 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.1 | 1.4 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.1 | 1.7 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.1 | 1.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.6 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.1 | 0.3 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.1 | 1.2 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 2.7 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.1 | 0.3 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.1 | 0.6 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 0.5 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 1.2 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.1 | 1.4 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 0.7 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 1.5 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.1 | 0.3 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.1 | 1.1 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.1 | 0.4 | GO:0036480 | neuron intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0036480) regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903376) positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 | 2.5 | GO:0007129 | synapsis(GO:0007129) |
| 0.1 | 0.7 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 | 0.2 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.1 | 1.3 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.1 | 0.8 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.1 | 1.1 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.1 | 0.2 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.1 | 5.0 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.1 | 0.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.5 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.8 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 0.2 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.1 | 1.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.2 | GO:0032415 | regulation of sodium:proton antiporter activity(GO:0032415) positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.1 | 0.3 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.1 | 0.2 | GO:1903406 | regulation of sodium:potassium-exchanging ATPase activity(GO:1903406) |
| 0.1 | 0.6 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.1 | 0.7 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) regulation of glucose import in response to insulin stimulus(GO:2001273) |
| 0.1 | 0.7 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 0.8 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 7.1 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.1 | 1.2 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.1 | 3.6 | GO:0043507 | positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 | 2.5 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.1 | 2.6 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.1 | 1.0 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.7 | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) |
| 0.1 | 0.5 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.1 | 2.4 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 0.7 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 1.3 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 0.9 | GO:2000780 | negative regulation of DNA repair(GO:0045738) negative regulation of double-strand break repair(GO:2000780) |
| 0.1 | 0.7 | GO:0000054 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.1 | 1.0 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.1 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 | 0.1 | GO:0050912 | detection of chemical stimulus involved in sensory perception(GO:0050907) detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.1 | 2.2 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.1 | 1.0 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 1.6 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.1 | 0.4 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 | 0.4 | GO:0090205 | positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 | 1.3 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.4 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.8 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 1.2 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.0 | 0.1 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.6 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.0 | 0.9 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.4 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.6 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 1.0 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 0.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.0 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.3 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.2 | GO:0090282 | trophectodermal cellular morphogenesis(GO:0001831) trophectodermal cell proliferation(GO:0001834) positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) positive regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0090282) |
| 0.0 | 0.5 | GO:0060441 | epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.2 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 1.4 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 1.0 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 | 1.1 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 1.2 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.2 | GO:2000110 | protein sialylation(GO:1990743) negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.0 | 2.3 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.1 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.1 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 2.1 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.3 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.6 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.8 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.4 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.4 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 1.1 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 0.1 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 | 0.4 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.4 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.7 | GO:0007623 | circadian rhythm(GO:0007623) |
| 0.0 | 0.2 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.1 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 1.9 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.8 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.2 | GO:1903351 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.0 | 0.3 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 0.2 | GO:0002828 | regulation of type 2 immune response(GO:0002828) |
| 0.0 | 0.3 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 1.1 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 1.2 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.3 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 | 0.1 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 1.9 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.4 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.2 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.1 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.0 | 0.3 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.0 | 0.8 | GO:0045931 | positive regulation of mitotic cell cycle(GO:0045931) |
| 0.0 | 0.0 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.3 | GO:0048255 | mRNA stabilization(GO:0048255) |
| 0.0 | 0.1 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 8.7 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 1.9 | 5.8 | GO:0071953 | elastic fiber(GO:0071953) |
| 1.6 | 6.3 | GO:0014802 | terminal cisterna(GO:0014802) |
| 1.2 | 6.2 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 1.0 | 5.9 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.9 | 2.8 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.9 | 2.7 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.9 | 6.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.8 | 5.0 | GO:0043256 | laminin complex(GO:0043256) |
| 0.7 | 2.2 | GO:0030934 | collagen type XII trimer(GO:0005595) anchoring collagen complex(GO:0030934) |
| 0.7 | 2.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.7 | 2.7 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.6 | 2.4 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.6 | 3.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.5 | 4.7 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.5 | 6.6 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.5 | 1.5 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.5 | 5.7 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.5 | 7.6 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.4 | 1.3 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.4 | 11.9 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.4 | 2.6 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.4 | 6.6 | GO:0005605 | basal lamina(GO:0005605) |
| 0.4 | 1.6 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.4 | 2.7 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.3 | 5.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.3 | 2.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.3 | 3.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.3 | 4.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 3.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.3 | 2.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.3 | 3.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.3 | 1.4 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.3 | 1.3 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.3 | 0.8 | GO:0044753 | amphisome(GO:0044753) |
| 0.3 | 6.5 | GO:0043657 | host(GO:0018995) host cell part(GO:0033643) host cell(GO:0043657) |
| 0.2 | 0.5 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.2 | 1.0 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.2 | 1.2 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.2 | 3.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.2 | 1.4 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.2 | 2.7 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 7.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.2 | 0.9 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.2 | 0.8 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.2 | 9.0 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 1.0 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.2 | 0.8 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.2 | 2.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.2 | 1.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.2 | 2.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.2 | 2.8 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.2 | 1.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.2 | 1.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 8.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.2 | 0.7 | GO:0032021 | NELF complex(GO:0032021) |
| 0.2 | 4.1 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 0.2 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.2 | 1.3 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.2 | 1.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.2 | 0.6 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.2 | 5.0 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 3.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 1.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.4 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 0.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 0.8 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 1.1 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 14.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 0.5 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 4.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 2.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.4 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.6 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 1.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 1.4 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 1.8 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 1.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.6 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 43.3 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 0.4 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 5.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 3.1 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 0.4 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 8.9 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.1 | 0.6 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.9 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 17.3 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 4.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.4 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 6.8 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 5.1 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 1.1 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 0.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 7.1 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 4.3 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 4.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 1.1 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 1.1 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 7.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 1.0 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 2.2 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.1 | 1.5 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 55.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 0.8 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 3.2 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 0.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.6 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.5 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 8.8 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.1 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.5 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 12.6 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 4.1 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 1.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.2 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.8 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 1.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 1.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.2 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 7.3 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.5 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.8 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.0 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.4 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.4 | GO:0030990 | intraciliary transport particle(GO:0030990) |
| 0.0 | 0.5 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.3 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.4 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.1 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.4 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.4 | GO:0031519 | PcG protein complex(GO:0031519) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 19.0 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 2.9 | 11.7 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 2.3 | 9.3 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 2.2 | 8.6 | GO:0008142 | oxysterol binding(GO:0008142) |
| 2.1 | 19.3 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 2.1 | 8.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 2.1 | 6.2 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 1.9 | 7.5 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 1.7 | 6.6 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 1.5 | 4.6 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 1.5 | 6.0 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 1.4 | 6.9 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 1.3 | 10.5 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 1.2 | 3.7 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 1.1 | 5.5 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 1.1 | 3.3 | GO:0042936 | dipeptide transporter activity(GO:0042936) |
| 1.0 | 3.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 1.0 | 3.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.9 | 5.5 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.9 | 3.6 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.9 | 11.3 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.9 | 3.4 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.8 | 2.4 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.8 | 8.8 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.8 | 7.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.8 | 18.4 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.6 | 3.1 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.6 | 4.9 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.6 | 6.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.6 | 1.8 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.6 | 1.7 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.6 | 1.7 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) hyaluronan synthase activity(GO:0050501) |
| 0.6 | 12.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.5 | 3.8 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.5 | 1.6 | GO:0019966 | C-X-C chemokine binding(GO:0019958) interleukin-1 binding(GO:0019966) tumor necrosis factor binding(GO:0043120) |
| 0.5 | 3.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.5 | 6.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.5 | 2.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.5 | 6.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.5 | 1.5 | GO:0032139 | DNA polymerase processivity factor activity(GO:0030337) dinucleotide insertion or deletion binding(GO:0032139) |
| 0.5 | 1.5 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.5 | 4.7 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.5 | 5.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.5 | 4.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.4 | 2.2 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.4 | 1.7 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.4 | 4.7 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.4 | 2.8 | GO:0036122 | BMP binding(GO:0036122) |
| 0.4 | 1.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.4 | 6.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.4 | 1.6 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.4 | 3.9 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.4 | 4.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.4 | 14.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.4 | 1.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.4 | 7.1 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.4 | 2.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.3 | 1.0 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.3 | 3.0 | GO:0032564 | dATP binding(GO:0032564) |
| 0.3 | 1.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.3 | 0.7 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.3 | 2.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.3 | 1.0 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.3 | 0.9 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.3 | 0.9 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.3 | 4.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.3 | 4.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.3 | 1.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.3 | 3.8 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.3 | 1.8 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.3 | 6.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.3 | 0.9 | GO:0004686 | elongation factor-2 kinase activity(GO:0004686) |
| 0.3 | 2.6 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.3 | 3.7 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.3 | 1.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.3 | 2.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.3 | 2.3 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.3 | 0.8 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.3 | 2.0 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.3 | 1.9 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.3 | 4.6 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.3 | 0.3 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.3 | 6.5 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.3 | 1.6 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.3 | 5.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.3 | 2.9 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.3 | 1.0 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.3 | 0.8 | GO:0004673 | phosphorelay sensor kinase activity(GO:0000155) protein histidine kinase activity(GO:0004673) |
| 0.3 | 2.9 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.3 | 0.8 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.3 | 1.5 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.3 | 1.3 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.3 | 2.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 1.5 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.2 | 1.0 | GO:0008948 | oxaloacetate decarboxylase activity(GO:0008948) |
| 0.2 | 0.5 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.2 | 8.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 1.4 | GO:0043426 | MRF binding(GO:0043426) |
| 0.2 | 1.2 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.2 | 3.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.2 | 1.4 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.2 | 4.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 1.4 | GO:0070728 | leucine binding(GO:0070728) |
| 0.2 | 2.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 0.7 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.2 | 0.9 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.2 | 0.8 | GO:0004104 | cholinesterase activity(GO:0004104) choline binding(GO:0033265) |
| 0.2 | 1.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.2 | 2.8 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.2 | 0.6 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.2 | 0.8 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.2 | 0.6 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.2 | 0.5 | GO:0019002 | GMP binding(GO:0019002) |
| 0.2 | 0.7 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.2 | 0.5 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.2 | 2.6 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.2 | 1.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.2 | 1.0 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.2 | 2.6 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.2 | 1.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.2 | 6.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.2 | 3.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 4.0 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.2 | 1.1 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.2 | 0.5 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.2 | 0.9 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 2.7 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.4 | GO:0017084 | glutamate 5-kinase activity(GO:0004349) glutamate-5-semialdehyde dehydrogenase activity(GO:0004350) delta1-pyrroline-5-carboxylate synthetase activity(GO:0017084) amino acid kinase activity(GO:0019202) |
| 0.1 | 1.4 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 9.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.9 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.1 | 1.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 1.7 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 1.4 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 0.7 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.3 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 1.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 1.2 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 0.3 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 3.8 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 0.8 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 1.7 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 1.3 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.1 | 2.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.6 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.1 | 11.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 3.5 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 0.4 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.1 | 3.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.6 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 0.6 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 4.1 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.1 | 0.2 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.1 | 0.2 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.1 | 2.6 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.1 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.1 | 1.0 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.1 | 0.6 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.1 | 1.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 2.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.6 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 0.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.9 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.1 | 2.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 2.9 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 1.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 5.0 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 1.1 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 0.1 | 1.2 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 0.5 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 1.0 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 0.5 | GO:0050694 | galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 1.3 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 0.8 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 3.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.4 | GO:0048029 | monosaccharide binding(GO:0048029) |
| 0.1 | 3.7 | GO:0020037 | heme binding(GO:0020037) |
| 0.1 | 0.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.6 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.1 | 0.8 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 1.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 0.2 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.1 | 0.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 1.5 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 0.7 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 7.2 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.1 | 2.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.7 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 1.0 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 1.8 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.1 | 0.4 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.1 | 0.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 6.9 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 1.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.9 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 1.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 1.0 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.6 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 1.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 3.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.9 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.3 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 8.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.4 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 0.2 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.6 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 1.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.7 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 1.9 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.2 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 0.6 | GO:0015149 | glucose transmembrane transporter activity(GO:0005355) hexose transmembrane transporter activity(GO:0015149) |
| 0.0 | 0.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 3.0 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 1.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 1.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.5 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.7 | GO:0030546 | receptor activator activity(GO:0030546) |
| 0.0 | 3.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.1 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 1.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 2.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 1.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.3 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.3 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.7 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.8 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.4 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.3 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.2 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 1.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.5 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 2.1 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.2 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 11.7 | PID_INTEGRIN2_PATHWAY | Beta2 integrin cell surface interactions |
| 0.7 | 10.3 | ST_JAK_STAT_PATHWAY | Jak-STAT Pathway |
| 0.6 | 5.0 | PID_INTEGRIN4_PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.5 | 6.4 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.4 | 2.6 | ST_STAT3_PATHWAY | STAT3 Pathway |
| 0.4 | 2.9 | PID_SHP2_PATHWAY | SHP2 signaling |
| 0.4 | 7.2 | PID_TCR_RAS_PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.4 | 17.9 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.3 | 2.0 | ST_PAC1_RECEPTOR_PATHWAY | PAC1 Receptor Pathway |
| 0.3 | 4.6 | NABA_BASEMENT_MEMBRANES | Genes encoding structural components of basement membranes |
| 0.3 | 16.5 | PID_INTEGRIN1_PATHWAY | Beta1 integrin cell surface interactions |
| 0.3 | 14.1 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.3 | 8.8 | PID_INSULIN_GLUCOSE_PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 27.5 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 10.7 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.2 | 6.7 | PID_PDGFRA_PATHWAY | PDGFR-alpha signaling pathway |
| 0.2 | 13.6 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.2 | 4.4 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 6.5 | PID_ER_NONGENOMIC_PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.2 | 25.2 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 12.2 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 7.6 | PID_HES_HEY_PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 3.4 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.1 | 5.3 | PID_ILK_PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 5.8 | PID_FAK_PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 1.1 | PID_VEGF_VEGFR_PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 3.5 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 0.8 | PID_ATF2_PATHWAY | ATF-2 transcription factor network |
| 0.1 | 3.9 | PID_ATR_PATHWAY | ATR signaling pathway |
| 0.1 | 1.3 | PID_ALPHA_SYNUCLEIN_PATHWAY | Alpha-synuclein signaling |
| 0.1 | 2.6 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 2.1 | PID_GLYPICAN_1PATHWAY | Glypican 1 network |
| 0.1 | 3.2 | ST_GAQ_PATHWAY | G alpha q Pathway |
| 0.1 | 1.4 | PID_HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 3.3 | PID_CD8_TCR_PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 0.3 | PID_LYMPH_ANGIOGENESIS_PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 2.0 | PID_EPHA_FWDPATHWAY | EPHA forward signaling |
| 0.1 | 1.5 | PID_P38_GAMMA_DELTA_PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 3.6 | PID_FGF_PATHWAY | FGF signaling pathway |
| 0.1 | 5.0 | PID_CDC42_PATHWAY | CDC42 signaling events |
| 0.1 | 1.8 | SIG_PIP3_SIGNALING_IN_B_LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 2.1 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.1 | 1.1 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.1 | 0.5 | PID_NFKAPPAB_CANONICAL_PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 1.4 | PID_A6B1_A6B4_INTEGRIN_PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 3.6 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 1.4 | PID_DNA_PK_PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 1.8 | PID_IL12_2PATHWAY | IL12-mediated signaling events |
| 0.1 | 0.6 | PID_S1P_META_PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 1.1 | PID_BARD1_PATHWAY | BARD1 signaling events |
| 0.1 | 2.5 | PID_NOTCH_PATHWAY | Notch signaling pathway |
| 0.1 | 0.5 | PID_RHODOPSIN_PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.4 | PID_PS1_PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 1.4 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.6 | PID_MET_PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 1.2 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.0 | 2.8 | WNT_SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.9 | ST_FAS_SIGNALING_PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.4 | PID_TNF_PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.1 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.0 | 0.4 | PID_ANGIOPOIETIN_RECEPTOR_PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.5 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.5 | PID_KIT_PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 2.4 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | PID_P53_REGULATION_PATHWAY | p53 pathway |
| 0.0 | 0.3 | PID_FANCONI_PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.1 | ST_ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.0 | ST_P38_MAPK_PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.2 | PID_RETINOIC_ACID_PATHWAY | Retinoic acid receptors-mediated signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 1.7 | REACTOME_SIGNALING_BY_NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.7 | 10.4 | REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.6 | 13.7 | REACTOME_IMMUNOREGULATORY_INTERACTIONS_BETWEEN_A_LYMPHOID_AND_A_NON_LYMPHOID_CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.6 | 6.7 | REACTOME_PROLACTIN_RECEPTOR_SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.6 | 17.7 | REACTOME_NUCLEAR_SIGNALING_BY_ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.5 | 1.0 | REACTOME_PLATELET_AGGREGATION_PLUG_FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.5 | 3.3 | REACTOME_SIGNALING_BY_NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.5 | 10.8 | REACTOME_HS_GAG_DEGRADATION | Genes involved in HS-GAG degradation |
| 0.5 | 4.7 | REACTOME_REMOVAL_OF_THE_FLAP_INTERMEDIATE_FROM_THE_C_STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.5 | 20.1 | REACTOME_GLYCOLYSIS | Genes involved in Glycolysis |
| 0.5 | 3.6 | REACTOME_TRANSPORT_OF_ORGANIC_ANIONS | Genes involved in Transport of organic anions |
| 0.4 | 6.9 | REACTOME_ACTIVATION_OF_THE_AP1_FAMILY_OF_TRANSCRIPTION_FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.4 | 7.2 | REACTOME_CONVERSION_FROM_APC_C_CDC20_TO_APC_C_CDH1_IN_LATE_ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.4 | 10.2 | REACTOME_REGULATION_OF_BETA_CELL_DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.3 | 2.8 | REACTOME_THE_ACTIVATION_OF_ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.3 | 2.8 | REACTOME_INHIBITION_OF_REPLICATION_INITIATION_OF_DAMAGED_DNA_BY_RB1_E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.3 | 3.4 | REACTOME_MRNA_DECAY_BY_3_TO_5_EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.3 | 18.7 | REACTOME_AMYLOIDS | Genes involved in Amyloids |
| 0.3 | 5.3 | REACTOME_SEMA3A_PLEXIN_REPULSION_SIGNALING_BY_INHIBITING_INTEGRIN_ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.3 | 9.3 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.3 | 3.5 | REACTOME_FGFR2C_LIGAND_BINDING_AND_ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.3 | 7.7 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.3 | 4.9 | REACTOME_APOPTOSIS_INDUCED_DNA_FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.3 | 8.0 | REACTOME_ENOS_ACTIVATION_AND_REGULATION | Genes involved in eNOS activation and regulation |
| 0.3 | 1.7 | REACTOME_ELEVATION_OF_CYTOSOLIC_CA2_LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.3 | 2.2 | REACTOME_HYALURONAN_UPTAKE_AND_DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.3 | 5.7 | REACTOME_CASPASE_MEDIATED_CLEAVAGE_OF_CYTOSKELETAL_PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.3 | 2.6 | REACTOME_INTRINSIC_PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 15.0 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.2 | 5.0 | REACTOME_GLUTATHIONE_CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 2.6 | REACTOME_IL_6_SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.2 | 3.4 | REACTOME_AKT_PHOSPHORYLATES_TARGETS_IN_THE_CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.2 | 3.7 | REACTOME_PRE_NOTCH_PROCESSING_IN_GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 14.5 | REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.2 | 2.3 | REACTOME_ASSOCIATION_OF_LICENSING_FACTORS_WITH_THE_PRE_REPLICATIVE_COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.2 | 2.3 | REACTOME_AMINE_DERIVED_HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 2.2 | REACTOME_NOTCH_HLH_TRANSCRIPTION_PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.2 | 2.5 | REACTOME_G1_S_SPECIFIC_TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.2 | 1.2 | REACTOME_PRE_NOTCH_TRANSCRIPTION_AND_TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.2 | 1.5 | REACTOME_G2_M_DNA_DAMAGE_CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.2 | 2.7 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 0.2 | 10.3 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 0.3 | REACTOME_CDC6_ASSOCIATION_WITH_THE_ORC_ORIGIN_COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 3.1 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.1 | 1.6 | REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 5.9 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 0.8 | REACTOME_CS_DS_DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 2.5 | REACTOME_CELL_EXTRACELLULAR_MATRIX_INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.4 | REACTOME_OXYGEN_DEPENDENT_PROLINE_HYDROXYLATION_OF_HYPOXIA_INDUCIBLE_FACTOR_ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 2.1 | REACTOME_METABOLISM_OF_PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.0 | REACTOME_BASE_FREE_SUGAR_PHOSPHATE_REMOVAL_VIA_THE_SINGLE_NUCLEOTIDE_REPLACEMENT_PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 0.7 | REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 6.9 | REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.9 | REACTOME_EFFECTS_OF_PIP2_HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 0.6 | REACTOME_FACILITATIVE_NA_INDEPENDENT_GLUCOSE_TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.9 | REACTOME_DEPOSITION_OF_NEW_CENPA_CONTAINING_NUCLEOSOMES_AT_THE_CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.2 | REACTOME_MTORC1_MEDIATED_SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 3.2 | REACTOME_GLUCOSE_TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.2 | REACTOME_PURINE_SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.1 | REACTOME_GLYCOGEN_BREAKDOWN_GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.8 | REACTOME_GLYCOSPHINGOLIPID_METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 0.4 | REACTOME_ADVANCED_GLYCOSYLATION_ENDPRODUCT_RECEPTOR_SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 1.5 | REACTOME_TRAFFICKING_OF_GLUR2_CONTAINING_AMPA_RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 0.6 | REACTOME_GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.2 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.3 | REACTOME_ALPHA_LINOLENIC_ACID_ALA_METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 0.7 | REACTOME_HDL_MEDIATED_LIPID_TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.9 | REACTOME_TRANSPORT_OF_VITAMINS_NUCLEOSIDES_AND_RELATED_MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 2.4 | REACTOME_RNA_POL_III_TRANSCRIPTION | Genes involved in RNA Polymerase III Transcription |
| 0.1 | 3.8 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 4.0 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 3.4 | REACTOME_TRANS_GOLGI_NETWORK_VESICLE_BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 3.3 | REACTOME_MHC_CLASS_II_ANTIGEN_PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.8 | REACTOME_FORMATION_OF_ATP_BY_CHEMIOSMOTIC_COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.9 | REACTOME_AMINE_COMPOUND_SLC_TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.5 | REACTOME_GAP_JUNCTION_DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 2.3 | REACTOME_AMINO_ACID_AND_OLIGOPEPTIDE_SLC_TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.5 | REACTOME_VEGF_LIGAND_RECEPTOR_INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME_DOWNREGULATION_OF_ERBB2_ERBB3_SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.1 | REACTOME_TIGHT_JUNCTION_INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.5 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_GOLGI_MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 1.6 | REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX_AND_SUBSEQUENTLY_THE_43S_COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.4 | REACTOME_BASIGIN_INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.7 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.6 | REACTOME_KERATAN_SULFATE_KERATIN_METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.0 | 0.7 | REACTOME_MITOCHONDRIAL_TRNA_AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.7 | REACTOME_CITRIC_ACID_CYCLE_TCA_CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.6 | REACTOME_RESPONSE_TO_ELEVATED_PLATELET_CYTOSOLIC_CA2_ | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.9 | REACTOME_ANTIVIRAL_MECHANISM_BY_IFN_STIMULATED_GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.4 | REACTOME_TIE2_SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.5 | REACTOME_NEGATIVE_REGULATORS_OF_RIG_I_MDA5_SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.2 | REACTOME_P2Y_RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME_SYNTHESIS_OF_SUBSTRATES_IN_N_GLYCAN_BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME_TERMINATION_OF_O_GLYCAN_BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.4 | REACTOME_MEIOTIC_RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.5 | REACTOME_PEPTIDE_CHAIN_ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME_SYNTHESIS_SECRETION_AND_DEACYLATION_OF_GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.3 | REACTOME_CIRCADIAN_REPRESSION_OF_EXPRESSION_BY_REV_ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.3 | REACTOME_EXTENSION_OF_TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 1.6 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.1 | REACTOME_RECYCLING_PATHWAY_OF_L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.1 | REACTOME_ACTIVATION_OF_GENES_BY_ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.2 | REACTOME_VITAMIN_B5_PANTOTHENATE_METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.2 | REACTOME_BIOSYNTHESIS_OF_THE_N_GLYCAN_PRECURSOR_DOLICHOL_LIPID_LINKED_OLIGOSACCHARIDE_LLO_AND_TRANSFER_TO_A_NASCENT_PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.2 | REACTOME_SRP_DEPENDENT_COTRANSLATIONAL_PROTEIN_TARGETING_TO_MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |