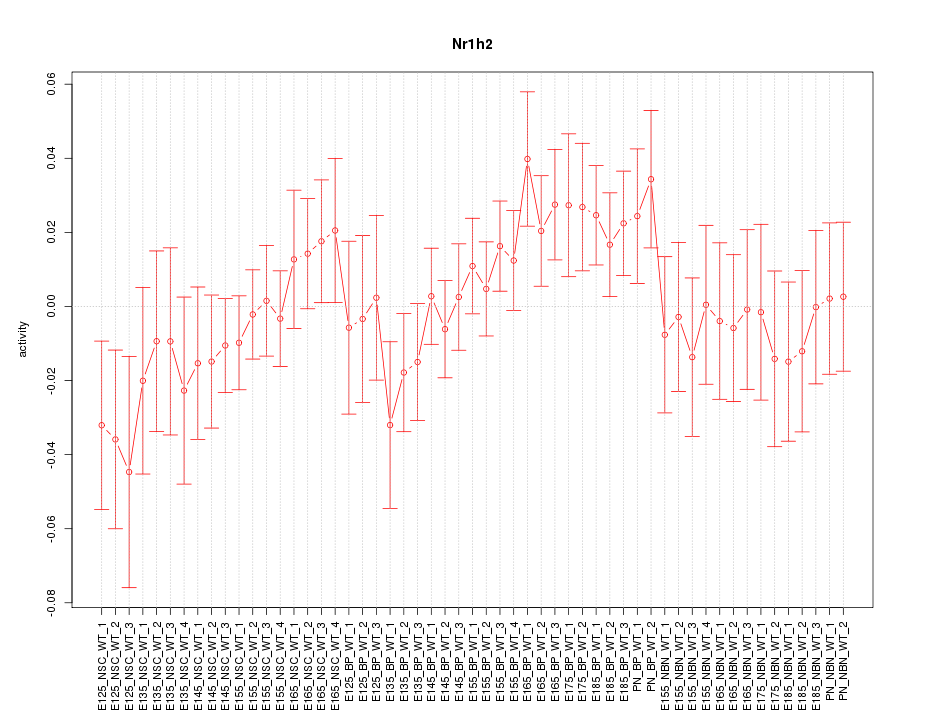
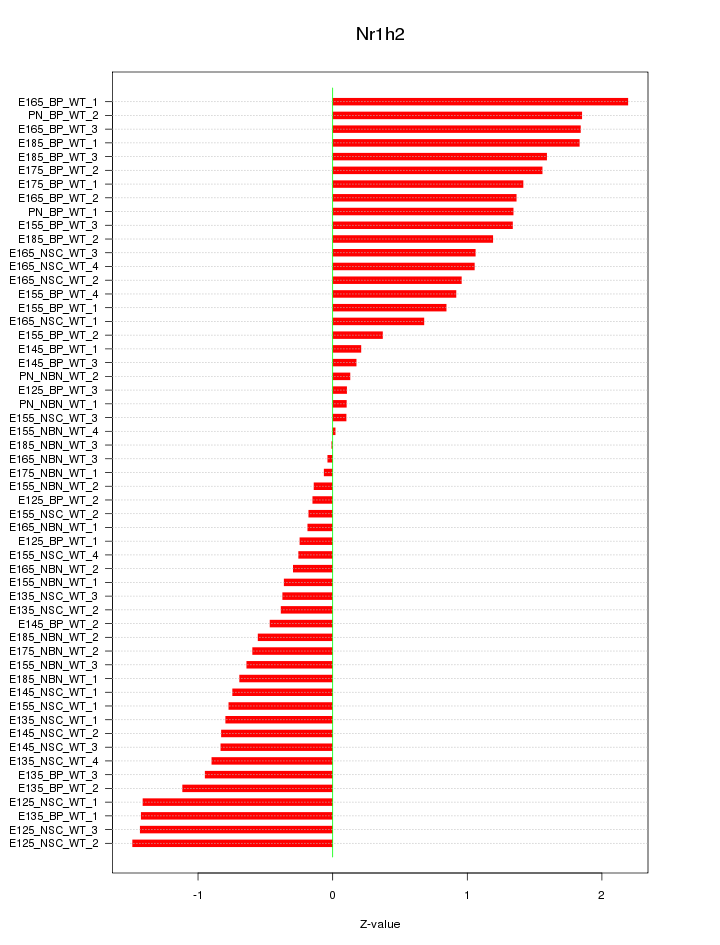
Motif ID: Nr1h2
Z-value: 0.965
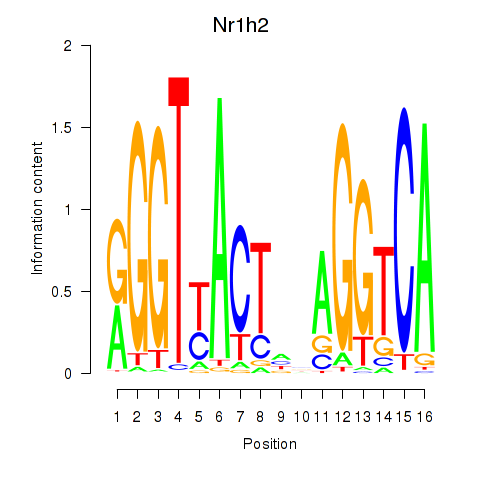
Transcription factors associated with Nr1h2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nr1h2 | ENSMUSG00000060601.6 | Nr1h2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr1h2 | mm10_v2_chr7_-_44553901_44553955 | 0.48 | 2.0e-04 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 11.7 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) platelet dense granule organization(GO:0060155) |
| 1.8 | 7.0 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.7 | 2.1 | GO:0014891 | striated muscle atrophy(GO:0014891) |
| 0.6 | 3.5 | GO:0032196 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) transposition(GO:0032196) |
| 0.6 | 1.7 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.6 | 4.5 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.6 | 3.4 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.5 | 3.6 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.5 | 1.4 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.5 | 2.3 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.5 | 1.4 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.4 | 1.8 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.4 | 1.7 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.3 | 7.4 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.3 | 0.6 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.3 | 3.7 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.3 | 2.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.2 | 0.7 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.2 | 0.9 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.2 | 1.8 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.2 | 1.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 | 9.6 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.2 | 0.6 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.2 | 0.9 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.2 | 0.7 | GO:0006558 | L-phenylalanine metabolic process(GO:0006558) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) |
| 0.2 | 1.5 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.2 | 1.6 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 3.2 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 | 3.4 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.2 | 1.0 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.2 | 1.6 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.2 | 3.1 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 2.6 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.1 | 0.8 | GO:0010730 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.1 | 0.5 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.1 | 0.7 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.1 | 1.0 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 1.6 | GO:0072112 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 | 1.1 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 0.4 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 | 2.2 | GO:0009988 | cell-cell recognition(GO:0009988) |
| 0.1 | 1.3 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.1 | 2.9 | GO:0060487 | lung epithelial cell differentiation(GO:0060487) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.7 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.4 | GO:1900194 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.5 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 0.4 | GO:0000022 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 1.8 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 1.0 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 3.7 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.5 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.0 | 0.6 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 1.9 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 2.3 | GO:0006073 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 0.1 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.0 | 3.3 | GO:0000398 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.4 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 | 0.1 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.4 | GO:1901998 | toxin transport(GO:1901998) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 11.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.4 | 2.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.3 | 2.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.3 | 1.0 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.2 | 1.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 2.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.2 | 1.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 0.8 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 3.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.5 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 1.8 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 3.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 1.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.6 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 3.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 3.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 6.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 3.0 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.1 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 1.5 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 3.2 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.7 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 2.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.7 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 2.3 | 7.0 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.9 | 3.5 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.6 | 3.6 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.5 | 1.4 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.4 | 1.7 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.4 | 1.2 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.4 | 1.1 | GO:0046592 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.3 | 1.3 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.3 | 2.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.3 | 1.6 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.3 | 1.5 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.3 | 1.4 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.2 | 1.0 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.2 | 2.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.2 | 0.9 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.2 | 2.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.2 | 1.7 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.2 | 1.0 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 1.6 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 0.4 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 1.8 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 0.6 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 1.0 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 9.6 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 3.7 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 0.4 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.4 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.6 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 4.5 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 0.4 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.1 | 0.7 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.1 | 1.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.8 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 1.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.4 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 3.7 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 1.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 1.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 2.7 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 1.4 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 0.9 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 2.0 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 1.8 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 3.1 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 2.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 16.2 | PID_RXR_VDR_PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.3 | 2.1 | SA_FAS_SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 1.6 | PID_ANTHRAX_PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 3.0 | PID_HES_HEY_PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 3.1 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
| 0.0 | 1.2 | PID_INTEGRIN_A9B1_PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 2.6 | PID_CD8_TCR_PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 3.4 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.4 | PID_IL6_7_PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.0 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.6 | PID_MTOR_4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.3 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.0 | 0.4 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 11.7 | REACTOME_HDL_MEDIATED_LIPID_TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.4 | 2.6 | REACTOME_ELEVATION_OF_CYTOSOLIC_CA2_LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.2 | 1.4 | REACTOME_ENDOGENOUS_STEROLS | Genes involved in Endogenous sterols |
| 0.2 | 2.7 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PC | Genes involved in Acyl chain remodelling of PC |
| 0.2 | 2.1 | REACTOME_EXTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 5.0 | REACTOME_FATTY_ACYL_COA_BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.1 | 4.5 | REACTOME_RORA_ACTIVATES_CIRCADIAN_EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.1 | 2.3 | REACTOME_METABOLISM_OF_POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.3 | REACTOME_FACILITATIVE_NA_INDEPENDENT_GLUCOSE_TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 2.3 | REACTOME_GLYCOGEN_BREAKDOWN_GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.7 | REACTOME_TRAF6_MEDIATED_IRF7_ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 1.6 | REACTOME_JNK_C_JUN_KINASES_PHOSPHORYLATION_AND_ACTIVATION_MEDIATED_BY_ACTIVATED_HUMAN_TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 3.6 | REACTOME_AMINO_ACID_AND_OLIGOPEPTIDE_SLC_TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 1.5 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.3 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.7 | REACTOME_BASIGIN_INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.4 | REACTOME_CONVERSION_FROM_APC_C_CDC20_TO_APC_C_CDH1_IN_LATE_ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.4 | REACTOME_ACTIVATED_AMPK_STIMULATES_FATTY_ACID_OXIDATION_IN_MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 1.0 | REACTOME_HEPARAN_SULFATE_HEPARIN_HS_GAG_METABOLISM | Genes involved in Heparan sulfate/heparin (HS-GAG) metabolism |
| 0.0 | 0.9 | REACTOME_METABOLISM_OF_VITAMINS_AND_COFACTORS | Genes involved in Metabolism of vitamins and cofactors |