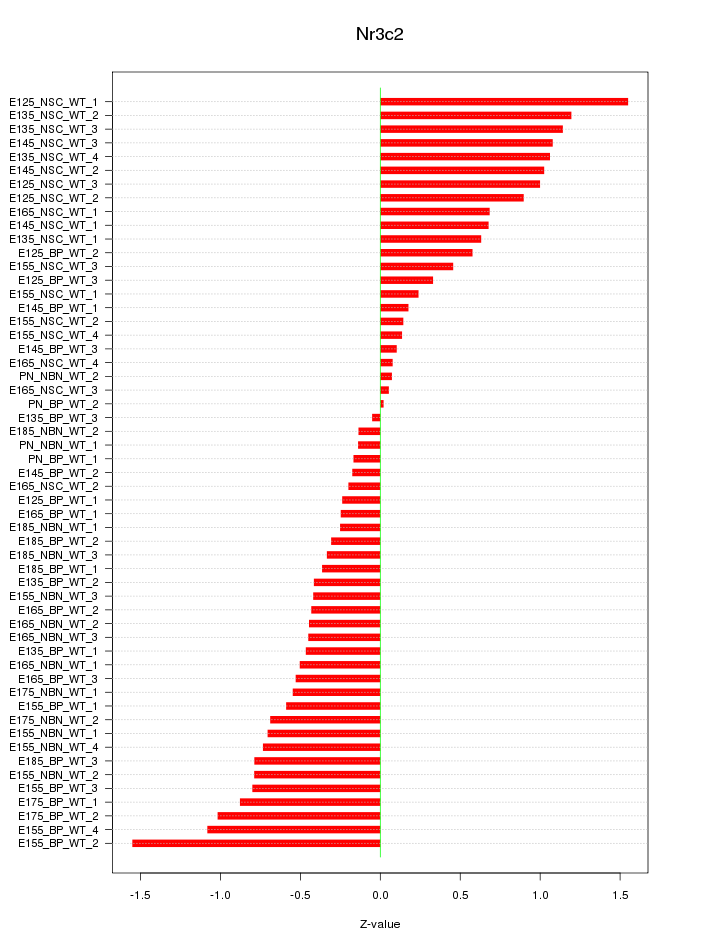
Motif ID: Nr3c2
Z-value: 0.663
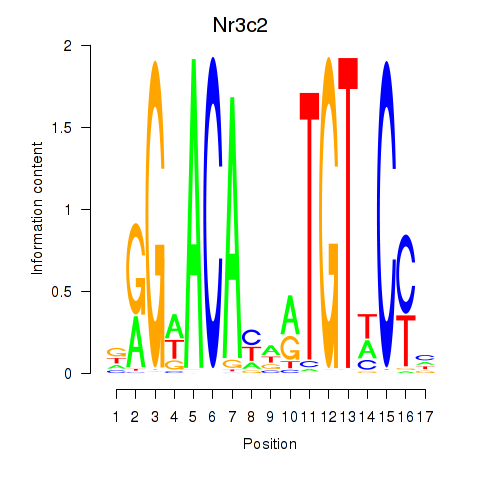
Transcription factors associated with Nr3c2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nr3c2 | ENSMUSG00000031618.7 | Nr3c2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr3c2 | mm10_v2_chr8_+_76902277_76902476 | -0.36 | 7.1e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 13.5 | GO:0060166 | olfactory pit development(GO:0060166) |
| 1.5 | 6.0 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 1.2 | 8.7 | GO:0010273 | detoxification of copper ion(GO:0010273) cellular response to zinc ion(GO:0071294) stress response to copper ion(GO:1990169) |
| 1.0 | 7.8 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.8 | 0.8 | GO:1904959 | regulation of electron carrier activity(GO:1904732) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.4 | 1.3 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.4 | 1.2 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.4 | 1.1 | GO:0042823 | pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.3 | 1.1 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.2 | 8.6 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.8 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 1.2 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.0 | 0.6 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 3.3 | GO:0008360 | regulation of cell shape(GO:0008360) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.0 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 1.3 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 0.8 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 12.8 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.2 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 7.8 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 4.3 | GO:0016607 | nuclear speck(GO:0016607) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 7.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.6 | 8.6 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.4 | 13.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.4 | 1.1 | GO:0031403 | lithium ion binding(GO:0031403) |
| 0.3 | 1.2 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.2 | 9.5 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 1.2 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 6.0 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 1.1 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 1.3 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 13.5 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 4.6 | PID_FANCONI_PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.8 | PID_FRA_PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.2 | PID_P73PATHWAY | p73 transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.6 | REACTOME_MITOCHONDRIAL_TRNA_AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 4.6 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.9 | REACTOME_INTERFERON_ALPHA_BETA_SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 3.3 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.1 | REACTOME_METABOLISM_OF_VITAMINS_AND_COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.2 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.6 | REACTOME_AMINO_ACID_TRANSPORT_ACROSS_THE_PLASMA_MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.8 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |