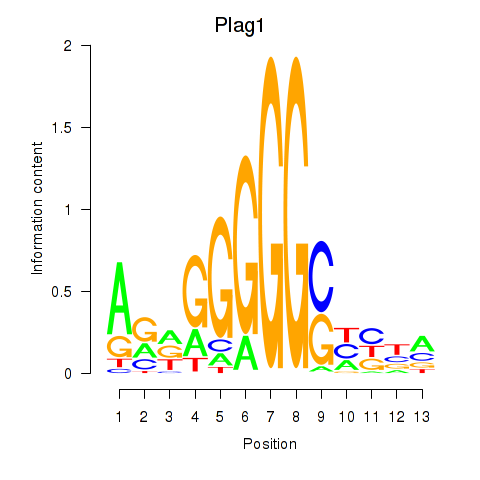
| 0.9 |
2.6 |
GO:0030827 |
negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.7 |
2.2 |
GO:0099548 |
drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.3 |
0.8 |
GO:1901079 |
positive regulation of relaxation of muscle(GO:1901079) |
| 0.2 |
0.7 |
GO:1903588 |
negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.2 |
1.6 |
GO:2001199 |
negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.2 |
0.6 |
GO:0016115 |
terpenoid catabolic process(GO:0016115) |
| 0.2 |
0.4 |
GO:1903587 |
regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.2 |
0.2 |
GO:0072309 |
mesenchymal stem cell maintenance involved in metanephric nephron morphogenesis(GO:0072309) |
| 0.1 |
0.8 |
GO:0032196 |
regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) transposition(GO:0032196) |
| 0.1 |
0.7 |
GO:1900748 |
positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 |
0.8 |
GO:0071630 |
nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.1 |
0.4 |
GO:0061144 |
alveolar secondary septum development(GO:0061144) |
| 0.1 |
0.4 |
GO:1905072 |
detection of oxygen(GO:0003032) cardiac jelly development(GO:1905072) |
| 0.1 |
0.5 |
GO:0070269 |
pyroptosis(GO:0070269) |
| 0.1 |
0.6 |
GO:0034441 |
plasma lipoprotein particle oxidation(GO:0034441) |
| 0.1 |
0.3 |
GO:0060489 |
orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.1 |
0.6 |
GO:1904995 |
negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.1 |
0.5 |
GO:0006680 |
glucosylceramide catabolic process(GO:0006680) |
| 0.1 |
0.8 |
GO:0035542 |
regulation of SNARE complex assembly(GO:0035542) |
| 0.1 |
0.4 |
GO:0003357 |
noradrenergic neuron differentiation(GO:0003357) |
| 0.1 |
0.6 |
GO:0032811 |
negative regulation of gamma-aminobutyric acid secretion(GO:0014053) negative regulation of epinephrine secretion(GO:0032811) |
| 0.1 |
0.3 |
GO:0060084 |
regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) synaptic transmission involved in micturition(GO:0060084) |
| 0.1 |
0.3 |
GO:0035790 |
platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.1 |
0.2 |
GO:0003195 |
tricuspid valve formation(GO:0003195) |
| 0.1 |
0.8 |
GO:0010459 |
negative regulation of heart rate(GO:0010459) |
| 0.1 |
0.3 |
GO:0048686 |
regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.1 |
0.2 |
GO:0007521 |
muscle cell fate determination(GO:0007521) |
| 0.1 |
0.4 |
GO:0061073 |
ciliary body morphogenesis(GO:0061073) |
| 0.1 |
0.8 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.1 |
0.4 |
GO:0061642 |
chemoattraction of axon(GO:0061642) |
| 0.1 |
1.7 |
GO:0090036 |
regulation of protein kinase C signaling(GO:0090036) |
| 0.1 |
0.4 |
GO:0007021 |
tubulin complex assembly(GO:0007021) |
| 0.1 |
0.3 |
GO:0003150 |
muscular septum morphogenesis(GO:0003150) |
| 0.1 |
0.3 |
GO:1904672 |
regulation of somatic stem cell population maintenance(GO:1904672) |
| 0.1 |
0.5 |
GO:0016185 |
synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.1 |
0.8 |
GO:0048843 |
negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.1 |
0.2 |
GO:0046340 |
diacylglycerol catabolic process(GO:0046340) |
| 0.1 |
1.0 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 |
0.2 |
GO:0060598 |
dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.1 |
0.1 |
GO:0038003 |
opioid receptor signaling pathway(GO:0038003) regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.1 |
0.3 |
GO:0071692 |
protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 0.1 |
0.2 |
GO:0032918 |
polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 |
0.2 |
GO:0001869 |
regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.1 |
0.5 |
GO:0060347 |
heart trabecula formation(GO:0060347) |
| 0.1 |
0.4 |
GO:0034316 |
negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 |
0.2 |
GO:0021933 |
radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 0.1 |
0.3 |
GO:0015803 |
branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 |
0.2 |
GO:0009298 |
GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 |
0.3 |
GO:0035022 |
positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 |
0.3 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 |
0.3 |
GO:0055091 |
phospholipid homeostasis(GO:0055091) |
| 0.1 |
0.5 |
GO:0010745 |
negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.1 |
0.4 |
GO:0045198 |
establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 |
0.3 |
GO:0048852 |
diencephalon morphogenesis(GO:0048852) |
| 0.1 |
0.5 |
GO:0048742 |
regulation of skeletal muscle fiber development(GO:0048742) |
| 0.1 |
0.3 |
GO:0071376 |
response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 |
0.3 |
GO:1905169 |
protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.0 |
0.2 |
GO:0060265 |
positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.0 |
0.5 |
GO:1900121 |
negative regulation of receptor binding(GO:1900121) |
| 0.0 |
0.1 |
GO:1902277 |
negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.0 |
0.2 |
GO:0070278 |
extracellular matrix constituent secretion(GO:0070278) |
| 0.0 |
0.1 |
GO:2000182 |
regulation of progesterone biosynthetic process(GO:2000182) |
| 0.0 |
0.0 |
GO:2000277 |
positive regulation of oxidative phosphorylation uncoupler activity(GO:2000277) |
| 0.0 |
0.3 |
GO:1903300 |
negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 |
0.2 |
GO:1903566 |
positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 |
0.6 |
GO:0048672 |
positive regulation of collateral sprouting(GO:0048672) |
| 0.0 |
0.4 |
GO:0032482 |
Rab protein signal transduction(GO:0032482) |
| 0.0 |
0.2 |
GO:0003192 |
mitral valve formation(GO:0003192) condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 |
0.1 |
GO:0070844 |
misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 |
2.4 |
GO:0021696 |
cerebellar cortex morphogenesis(GO:0021696) |
| 0.0 |
1.8 |
GO:0007218 |
neuropeptide signaling pathway(GO:0007218) |
| 0.0 |
0.2 |
GO:0097211 |
response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 |
0.2 |
GO:0003376 |
sphingosine-1-phosphate signaling pathway(GO:0003376) eye pigmentation(GO:0048069) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 |
1.1 |
GO:0033198 |
response to ATP(GO:0033198) |
| 0.0 |
0.1 |
GO:0098974 |
postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.0 |
0.2 |
GO:0035021 |
negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 |
0.7 |
GO:0033234 |
negative regulation of protein sumoylation(GO:0033234) |
| 0.0 |
0.2 |
GO:0045759 |
negative regulation of action potential(GO:0045759) |
| 0.0 |
0.0 |
GO:1904008 |
response to monosodium glutamate(GO:1904008) cellular response to monosodium glutamate(GO:1904009) |
| 0.0 |
0.1 |
GO:0016561 |
protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 |
0.0 |
GO:0060003 |
copper ion export(GO:0060003) |
| 0.0 |
0.3 |
GO:0071802 |
negative regulation of podosome assembly(GO:0071802) |
| 0.0 |
0.2 |
GO:0045078 |
positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 |
0.2 |
GO:0099525 |
presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 |
0.2 |
GO:1905150 |
regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 |
0.3 |
GO:0055003 |
cardiac myofibril assembly(GO:0055003) |
| 0.0 |
0.1 |
GO:0061428 |
negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 |
0.5 |
GO:0099628 |
receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.0 |
0.1 |
GO:1904529 |
regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.0 |
0.2 |
GO:0000189 |
MAPK import into nucleus(GO:0000189) regulation of Golgi inheritance(GO:0090170) |
| 0.0 |
0.1 |
GO:1902268 |
negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 |
0.1 |
GO:1902608 |
regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.0 |
0.1 |
GO:2000256 |
positive regulation of male germ cell proliferation(GO:2000256) |
| 0.0 |
0.3 |
GO:0001842 |
neural fold formation(GO:0001842) |
| 0.0 |
0.2 |
GO:0097473 |
cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.0 |
0.3 |
GO:0042989 |
sequestering of actin monomers(GO:0042989) |
| 0.0 |
0.1 |
GO:0021681 |
cerebellar granular layer development(GO:0021681) defecation(GO:0030421) |
| 0.0 |
0.1 |
GO:0072720 |
response to dithiothreitol(GO:0072720) |
| 0.0 |
0.4 |
GO:0035646 |
endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.0 |
0.1 |
GO:1901526 |
positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 |
0.2 |
GO:0072318 |
clathrin coat disassembly(GO:0072318) |
| 0.0 |
0.1 |
GO:0051582 |
positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.0 |
0.2 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.0 |
0.2 |
GO:0071985 |
multivesicular body sorting pathway(GO:0071985) |
| 0.0 |
0.1 |
GO:0071211 |
protein targeting to vacuole involved in autophagy(GO:0071211) |
| 0.0 |
0.2 |
GO:0021780 |
oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 |
0.1 |
GO:1990034 |
calcium ion export from cell(GO:1990034) |
| 0.0 |
0.1 |
GO:1902220 |
positive regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902220) |
| 0.0 |
0.1 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.0 |
0.5 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.0 |
0.1 |
GO:0019482 |
beta-alanine metabolic process(GO:0019482) |
| 0.0 |
0.1 |
GO:0031648 |
protein destabilization(GO:0031648) |
| 0.0 |
0.2 |
GO:0035754 |
B cell chemotaxis(GO:0035754) |
| 0.0 |
0.1 |
GO:0090063 |
positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 |
0.1 |
GO:0010726 |
positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.0 |
0.1 |
GO:0090327 |
negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.0 |
0.1 |
GO:1905167 |
positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.0 |
0.2 |
GO:0072321 |
chaperone-mediated protein transport(GO:0072321) |
| 0.0 |
0.2 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 |
0.2 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 |
0.1 |
GO:0021592 |
fourth ventricle development(GO:0021592) initiation of neural tube closure(GO:0021993) |
| 0.0 |
0.1 |
GO:0035984 |
response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.0 |
0.1 |
GO:0048105 |
establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.0 |
0.0 |
GO:0046619 |
optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 |
0.3 |
GO:0061577 |
calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) |
| 0.0 |
0.1 |
GO:0051697 |
protein delipidation(GO:0051697) |
| 0.0 |
0.0 |
GO:0015919 |
peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 |
0.1 |
GO:0019244 |
lactate biosynthetic process from pyruvate(GO:0019244) |
| 0.0 |
0.2 |
GO:0048102 |
autophagic cell death(GO:0048102) |
| 0.0 |
0.1 |
GO:0042373 |
vitamin K metabolic process(GO:0042373) |
| 0.0 |
0.2 |
GO:1990573 |
potassium ion import across plasma membrane(GO:1990573) |
| 0.0 |
0.1 |
GO:0021631 |
optic nerve morphogenesis(GO:0021631) regulation of circadian sleep/wake cycle, non-REM sleep(GO:0045188) |
| 0.0 |
0.1 |
GO:2000048 |
negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 |
0.2 |
GO:0045671 |
negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 |
0.4 |
GO:0007263 |
nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 |
0.2 |
GO:0006527 |
arginine catabolic process(GO:0006527) |
| 0.0 |
0.1 |
GO:0072015 |
glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 |
0.3 |
GO:0044804 |
nucleophagy(GO:0044804) |
| 0.0 |
0.1 |
GO:0042535 |
positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.0 |
0.0 |
GO:0046671 |
negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 |
0.2 |
GO:0035428 |
hexose transmembrane transport(GO:0035428) |
| 0.0 |
0.1 |
GO:0051902 |
negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.0 |
0.0 |
GO:0070358 |
actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 |
0.3 |
GO:0006995 |
cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 |
0.3 |
GO:0045579 |
positive regulation of B cell differentiation(GO:0045579) |
| 0.0 |
0.1 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 |
0.1 |
GO:0042590 |
antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 |
0.1 |
GO:2001184 |
regulation of interleukin-12 secretion(GO:2001182) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 |
0.1 |
GO:0036481 |
intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036481) |
| 0.0 |
0.1 |
GO:0071677 |
positive regulation of mononuclear cell migration(GO:0071677) |
| 0.0 |
0.2 |
GO:0032225 |
regulation of synaptic transmission, dopaminergic(GO:0032225) |
| 0.0 |
0.1 |
GO:0050965 |
detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 |
0.1 |
GO:0046368 |
GDP-L-fucose metabolic process(GO:0046368) |
| 0.0 |
0.1 |
GO:1903056 |
positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.0 |
0.1 |
GO:0060178 |
regulation of exocyst localization(GO:0060178) |
| 0.0 |
0.1 |
GO:1902998 |
macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 |
0.2 |
GO:0090315 |
negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 |
0.2 |
GO:0046339 |
diacylglycerol metabolic process(GO:0046339) |
| 0.0 |
0.1 |
GO:0097274 |
urea homeostasis(GO:0097274) |
| 0.0 |
0.2 |
GO:1903818 |
positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 |
0.1 |
GO:2000774 |
positive regulation of cellular senescence(GO:2000774) |
| 0.0 |
0.1 |
GO:0052490 |
suppression by virus of host apoptotic process(GO:0019050) negative regulation by symbiont of host apoptotic process(GO:0033668) modulation by virus of host apoptotic process(GO:0039526) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 0.0 |
0.3 |
GO:0051988 |
regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 |
0.1 |
GO:0017185 |
peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 |
0.1 |
GO:1902669 |
positive regulation of axon guidance(GO:1902669) |
| 0.0 |
0.3 |
GO:0010460 |
positive regulation of heart rate(GO:0010460) |
| 0.0 |
0.1 |
GO:0032495 |
response to muramyl dipeptide(GO:0032495) |
| 0.0 |
0.0 |
GO:0043379 |
rRNA export from nucleus(GO:0006407) memory T cell differentiation(GO:0043379) response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 |
0.0 |
GO:2000793 |
cell proliferation involved in heart valve development(GO:2000793) |
| 0.0 |
0.1 |
GO:2000773 |
negative regulation of cellular senescence(GO:2000773) |
| 0.0 |
0.1 |
GO:1903358 |
regulation of Golgi organization(GO:1903358) |
| 0.0 |
0.1 |
GO:0033540 |
fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 |
0.3 |
GO:0033169 |
histone H3-K9 demethylation(GO:0033169) |
| 0.0 |
0.1 |
GO:0042760 |
very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 |
0.0 |
GO:0070649 |
formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 |
0.0 |
GO:0048611 |
ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.0 |
0.1 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.0 |
0.1 |
GO:0016557 |
peroxisome membrane biogenesis(GO:0016557) |
| 0.0 |
0.2 |
GO:1990440 |
positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 |
0.1 |
GO:0098734 |
protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 |
0.2 |
GO:0042178 |
xenobiotic catabolic process(GO:0042178) |
| 0.0 |
0.2 |
GO:1900746 |
regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.0 |
1.5 |
GO:0010811 |
positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 |
0.0 |
GO:0016554 |
cytidine to uridine editing(GO:0016554) |
| 0.0 |
0.1 |
GO:0009405 |
pathogenesis(GO:0009405) |
| 0.0 |
0.1 |
GO:0031000 |
response to caffeine(GO:0031000) |
| 0.0 |
0.0 |
GO:0006546 |
glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 |
0.0 |
GO:2000138 |
positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.0 |
0.1 |
GO:0050651 |
dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 |
0.0 |
GO:1904580 |
regulation of intracellular mRNA localization(GO:1904580) |
| 0.0 |
0.2 |
GO:0006516 |
glycoprotein catabolic process(GO:0006516) |
| 0.0 |
0.1 |
GO:1901096 |
regulation of autophagosome maturation(GO:1901096) |
| 0.0 |
0.0 |
GO:1901675 |
negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 |
0.1 |
GO:2000786 |
positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 |
0.1 |
GO:0001956 |
positive regulation of neurotransmitter secretion(GO:0001956) positive regulation of neurotransmitter transport(GO:0051590) |
| 0.0 |
0.1 |
GO:0046548 |
retinal rod cell development(GO:0046548) |
| 0.0 |
0.1 |
GO:0071763 |
nuclear membrane organization(GO:0071763) |
| 0.0 |
0.3 |
GO:0045026 |
plasma membrane fusion(GO:0045026) |
| 0.0 |
0.2 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.0 |
0.2 |
GO:0035058 |
nonmotile primary cilium assembly(GO:0035058) regulation of nonmotile primary cilium assembly(GO:1902855) positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 |
0.0 |
GO:0015772 |
disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 |
0.2 |
GO:0048712 |
negative regulation of astrocyte differentiation(GO:0048712) |
| 0.0 |
0.0 |
GO:0021886 |
hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.0 |
0.1 |
GO:0070445 |
oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 |
0.1 |
GO:0009437 |
carnitine metabolic process(GO:0009437) |
| 0.0 |
0.3 |
GO:0035455 |
response to interferon-alpha(GO:0035455) |
| 0.0 |
0.4 |
GO:0050919 |
negative chemotaxis(GO:0050919) |
| 0.0 |
0.4 |
GO:0021591 |
ventricular system development(GO:0021591) |
| 0.0 |
0.4 |
GO:0060351 |
cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.0 |
0.1 |
GO:0071539 |
protein localization to centrosome(GO:0071539) |
| 0.0 |
0.1 |
GO:0033539 |
fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 |
0.2 |
GO:0003334 |
keratinocyte development(GO:0003334) |
| 0.0 |
0.3 |
GO:0040015 |
negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 |
0.0 |
GO:0000429 |
carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) carbon catabolite activation of transcription(GO:0045991) |
| 0.0 |
0.2 |
GO:2000311 |
regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 |
0.1 |
GO:0006682 |
galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 |
0.1 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 |
0.0 |
GO:0001830 |
trophectodermal cell fate commitment(GO:0001830) |