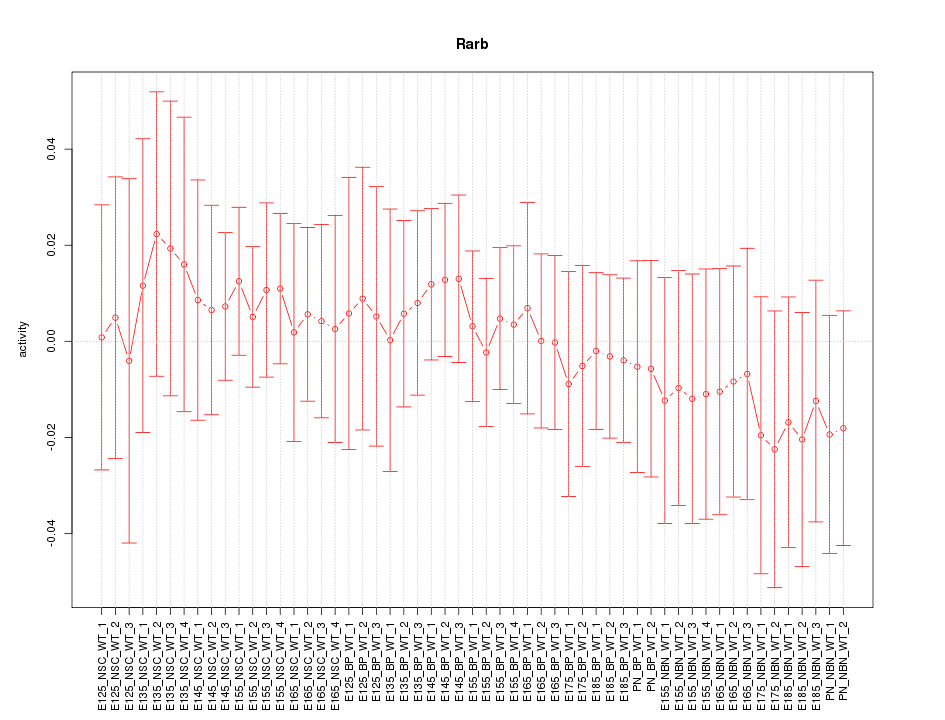
Motif ID: Rarb
Z-value: 0.451
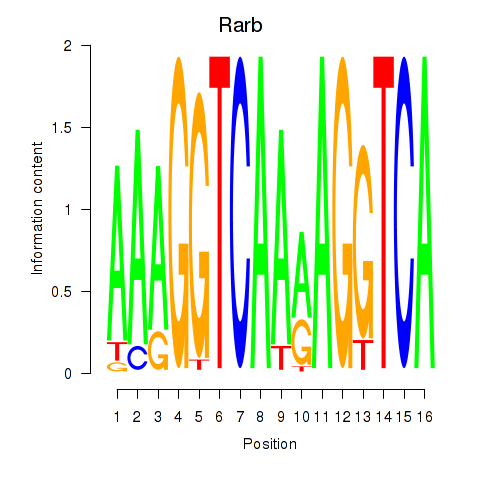
Transcription factors associated with Rarb:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Rarb | ENSMUSG00000017491.8 | Rarb |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rarb | mm10_v2_chr14_-_16575456_16575501 | -0.11 | 4.2e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.4 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.7 | 6.5 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.6 | 6.6 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.3 | 2.8 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.2 | 1.2 | GO:0098734 | protein depalmitoylation(GO:0002084) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) negative regulation of establishment of protein localization to plasma membrane(GO:0090005) macromolecule depalmitoylation(GO:0098734) |
| 0.2 | 0.6 | GO:1904959 | regulation of electron carrier activity(GO:1904732) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.1 | 5.5 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 2.9 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 4.2 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.0 | 0.6 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 1.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.2 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 1.3 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.9 | GO:0043966 | histone H3 acetylation(GO:0043966) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 0.6 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 6.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 5.4 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.1 | 5.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.1 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.9 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 4.2 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 6.6 | GO:0005667 | transcription factor complex(GO:0005667) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) C-X-C chemokine binding(GO:0019958) |
| 1.6 | 6.5 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 1.2 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.6 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 6.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 5.4 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 4.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.1 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.9 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 2.8 | GO:0003713 | transcription coactivator activity(GO:0003713) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.4 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.1 | 6.5 | PID_RAC1_PATHWAY | RAC1 signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.9 | REACTOME_RECRUITMENT_OF_NUMA_TO_MITOTIC_CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 5.4 | REACTOME_APC_CDC20_MEDIATED_DEGRADATION_OF_NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.2 | 2.8 | REACTOME_NOTCH_HLH_TRANSCRIPTION_PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.1 | REACTOME_MRNA_SPLICING_MINOR_PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.6 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |