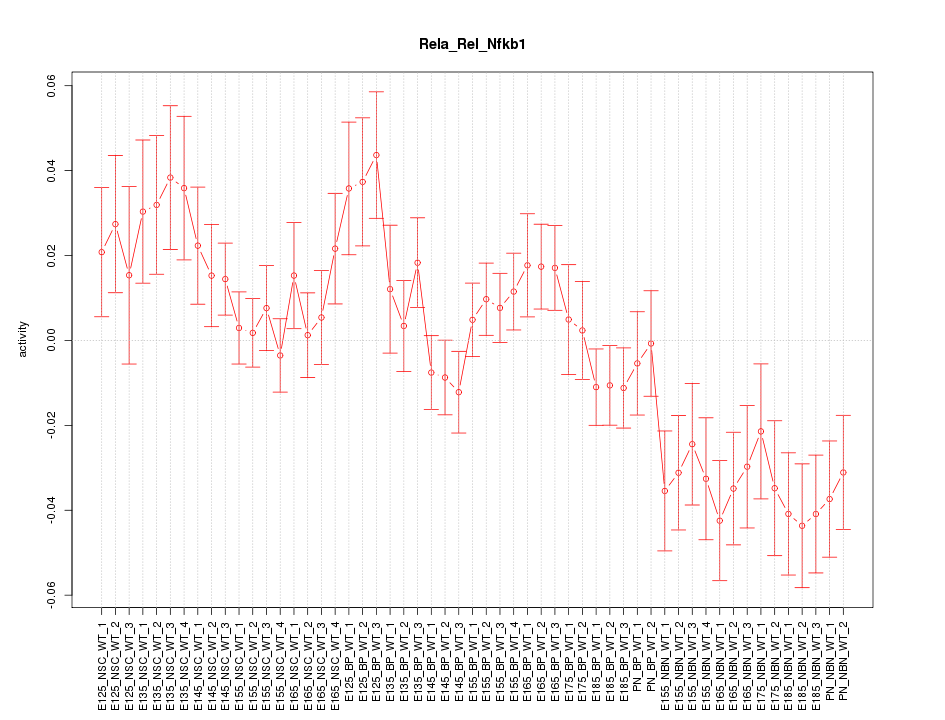
Motif ID: Rela_Rel_Nfkb1
Z-value: 1.723
Transcription factors associated with Rela_Rel_Nfkb1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nfkb1 | ENSMUSG00000028163.11 | Nfkb1 |
| Rel | ENSMUSG00000020275.8 | Rel |
| Rela | ENSMUSG00000024927.7 | Rela |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfkb1 | mm10_v2_chr3_-_135608221_135608290 | 0.72 | 8.3e-10 | Click! |
| Rela | mm10_v2_chr19_+_5637475_5637486 | 0.43 | 1.1e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.0 | 63.0 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 5.6 | 16.9 | GO:0097350 | neutrophil clearance(GO:0097350) |
| 4.2 | 12.6 | GO:0032916 | positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 3.6 | 10.9 | GO:0072554 | arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) blood vessel lumenization(GO:0072554) blood vessel endothelial cell fate specification(GO:0097101) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of heart induction(GO:1901321) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 2.8 | 8.5 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 2.7 | 8.2 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 2.0 | 12.2 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 2.0 | 6.1 | GO:0016115 | terpenoid catabolic process(GO:0016115) |
| 2.0 | 23.5 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 1.9 | 13.6 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 1.8 | 7.3 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 1.8 | 21.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 1.7 | 6.7 | GO:1903416 | negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) regulation of calcidiol 1-monooxygenase activity(GO:0060558) response to glycoside(GO:1903416) |
| 1.6 | 6.3 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 1.5 | 14.8 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 1.4 | 14.3 | GO:0000022 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 1.4 | 2.8 | GO:0033864 | positive regulation of NAD(P)H oxidase activity(GO:0033864) |
| 1.4 | 4.2 | GO:0090403 | regulation of purine nucleotide catabolic process(GO:0033121) negative regulation of purine nucleotide catabolic process(GO:0033122) oxidative stress-induced premature senescence(GO:0090403) |
| 1.4 | 7.0 | GO:0015671 | oxygen transport(GO:0015671) |
| 1.4 | 2.8 | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070432) |
| 1.4 | 2.7 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 1.3 | 20.9 | GO:0003334 | keratinocyte development(GO:0003334) |
| 1.3 | 3.9 | GO:1904170 | regulation of bleb assembly(GO:1904170) |
| 1.3 | 1.3 | GO:0033860 | regulation of NAD(P)H oxidase activity(GO:0033860) |
| 1.2 | 4.9 | GO:0030091 | protein repair(GO:0030091) |
| 1.2 | 3.7 | GO:0006116 | NADH oxidation(GO:0006116) |
| 1.2 | 7.0 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 1.2 | 1.2 | GO:0002678 | positive regulation of chronic inflammatory response(GO:0002678) |
| 1.1 | 4.5 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 1.1 | 8.8 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 1.1 | 4.4 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 1.1 | 3.3 | GO:0015938 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) acetyl-CoA catabolic process(GO:0046356) |
| 1.1 | 5.3 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 1.0 | 3.1 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 1.0 | 7.0 | GO:0007296 | vitellogenesis(GO:0007296) |
| 1.0 | 5.0 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 1.0 | 21.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 1.0 | 13.6 | GO:0002467 | germinal center formation(GO:0002467) |
| 1.0 | 4.8 | GO:2000152 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) regulation of ubiquitin-specific protease activity(GO:2000152) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.9 | 4.7 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.9 | 3.4 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.9 | 1.7 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.8 | 2.5 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.8 | 3.3 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.8 | 4.0 | GO:0090666 | telomere assembly(GO:0032202) scaRNA localization to Cajal body(GO:0090666) |
| 0.8 | 7.2 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.8 | 2.4 | GO:0060023 | soft palate development(GO:0060023) |
| 0.7 | 3.0 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.7 | 6.6 | GO:0042095 | interferon-gamma biosynthetic process(GO:0042095) |
| 0.7 | 2.2 | GO:1902460 | transforming growth factor beta activation(GO:0036363) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.7 | 4.2 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.7 | 9.4 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.7 | 4.7 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.7 | 2.0 | GO:0035934 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.6 | 3.2 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.6 | 3.2 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.6 | 10.1 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.6 | 1.8 | GO:0090135 | actin filament branching(GO:0090135) negative regulation of phospholipase C activity(GO:1900275) |
| 0.6 | 2.4 | GO:0032382 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 0.6 | 1.7 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.6 | 7.5 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.6 | 2.3 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.6 | 2.2 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.6 | 1.7 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.5 | 5.5 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.5 | 2.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) pyroptosis(GO:0070269) |
| 0.5 | 2.7 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.5 | 3.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.5 | 5.4 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.5 | 2.1 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.5 | 1.6 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.5 | 1.4 | GO:1901250 | regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.5 | 3.8 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.5 | 2.4 | GO:1905076 | regulation of interleukin-17 secretion(GO:1905076) |
| 0.5 | 5.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.4 | 3.9 | GO:2000254 | regulation of male germ cell proliferation(GO:2000254) |
| 0.4 | 8.2 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.4 | 6.7 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.4 | 2.1 | GO:1900153 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.4 | 1.2 | GO:0035622 | intrahepatic bile duct development(GO:0035622) cholangiocyte proliferation(GO:1990705) |
| 0.4 | 10.0 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.4 | 0.8 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.4 | 2.9 | GO:0048625 | embryonic hindgut morphogenesis(GO:0048619) myoblast fate commitment(GO:0048625) |
| 0.4 | 1.8 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.4 | 1.4 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.4 | 1.4 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.3 | 1.0 | GO:0033085 | negative regulation of T cell differentiation in thymus(GO:0033085) |
| 0.3 | 1.4 | GO:0006547 | histidine metabolic process(GO:0006547) |
| 0.3 | 1.4 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.3 | 3.4 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.3 | 1.0 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.3 | 6.1 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.3 | 1.9 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.3 | 1.3 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.3 | 2.8 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.3 | 2.7 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.3 | 2.7 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.3 | 2.6 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.3 | 1.2 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.3 | 0.9 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.3 | 1.7 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.3 | 2.2 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.3 | 0.8 | GO:0032621 | interleukin-18 production(GO:0032621) |
| 0.3 | 1.4 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.3 | 0.8 | GO:0030321 | transepithelial chloride transport(GO:0030321) transepithelial ammonium transport(GO:0070634) |
| 0.3 | 0.8 | GO:1904884 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.3 | 5.0 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.3 | 1.3 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.3 | 1.3 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.3 | 2.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.3 | 13.6 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.2 | 3.2 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.2 | 1.5 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.2 | 1.5 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.2 | 4.4 | GO:0045830 | positive regulation of isotype switching(GO:0045830) |
| 0.2 | 0.7 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.2 | 2.7 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.2 | 0.9 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.2 | 2.8 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.2 | 1.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.2 | 0.5 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.2 | 1.4 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.2 | 1.6 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.2 | 4.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.2 | 3.6 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.2 | 2.4 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.2 | 12.6 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.2 | 2.4 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.2 | 4.8 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.2 | 2.1 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.2 | 2.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.2 | 0.6 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.2 | 1.5 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.2 | 1.9 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.2 | 4.7 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.2 | 0.9 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.2 | 0.6 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.2 | 0.3 | GO:0071864 | positive regulation of cell proliferation in bone marrow(GO:0071864) |
| 0.2 | 1.0 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.2 | 1.8 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.2 | 0.3 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.2 | 0.6 | GO:0010896 | negative regulation of sequestering of triglyceride(GO:0010891) regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.2 | 1.4 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.2 | 1.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.2 | 1.8 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.2 | 0.6 | GO:1902340 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.1 | 1.6 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 0.9 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.1 | 0.7 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 1.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 3.8 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.1 | 2.7 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.1 | 0.9 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 | 3.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.3 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.1 | 4.7 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.4 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.1 | 3.8 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.2 | GO:1904245 | regulation of polynucleotide adenylyltransferase activity(GO:1904245) |
| 0.1 | 12.1 | GO:0007179 | transforming growth factor beta receptor signaling pathway(GO:0007179) |
| 0.1 | 0.4 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.1 | 2.0 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 0.5 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 2.2 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.1 | 0.6 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 0.8 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 2.2 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.1 | 1.1 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.1 | 1.3 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.1 | 0.9 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.6 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.1 | 3.4 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.1 | 0.8 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.1 | 1.2 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.1 | 2.0 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.1 | 2.6 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.3 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.1 | 0.4 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 3.3 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 0.8 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.1 | 3.7 | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) |
| 0.1 | 0.4 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.3 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.1 | 0.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.2 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.1 | 2.8 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.1 | 0.7 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 0.3 | GO:0070633 | transepithelial transport(GO:0070633) |
| 0.1 | 0.5 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 0.4 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.1 | 0.4 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 | 1.8 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 1.5 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.1 | 0.3 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.1 | 0.5 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.6 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 2.2 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 | 0.7 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 0.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 1.5 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 1.6 | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.1 | 0.1 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.1 | 1.7 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 4.3 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.1 | 1.0 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.1 | 0.7 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.1 | 0.3 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.1 | 1.1 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 0.2 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.1 | 2.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.4 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.0 | 0.2 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.9 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.6 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.0 | 2.3 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 4.2 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 0.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.5 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 2.3 | GO:0006959 | humoral immune response(GO:0006959) |
| 0.0 | 1.3 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.7 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.2 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 | 0.5 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.8 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.1 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 2.4 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.7 | GO:0051352 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) |
| 0.0 | 0.7 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.0 | 1.0 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.4 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 4.4 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.7 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.2 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 2.8 | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation(GO:0050731) |
| 0.0 | 1.5 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.8 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.1 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 1.4 | GO:0071774 | response to fibroblast growth factor(GO:0071774) |
| 0.0 | 0.6 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.8 | GO:0006505 | GPI anchor metabolic process(GO:0006505) GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.6 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.2 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.2 | GO:0042182 | ketone catabolic process(GO:0042182) |
| 0.0 | 0.1 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.0 | 0.1 | GO:0031274 | regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.4 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.7 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.1 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.1 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.0 | 0.7 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.2 | GO:0048538 | thymus development(GO:0048538) |
| 0.0 | 0.1 | GO:0044241 | lipid digestion(GO:0044241) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 18.9 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 2.2 | 10.9 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 1.9 | 11.2 | GO:0098536 | deuterosome(GO:0098536) |
| 1.8 | 5.4 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 1.6 | 8.2 | GO:0031523 | Myb complex(GO:0031523) |
| 1.5 | 4.5 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 1.2 | 5.0 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 1.2 | 4.9 | GO:0043293 | apoptosome(GO:0043293) |
| 0.9 | 3.7 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.9 | 4.4 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.9 | 7.0 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.9 | 5.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.8 | 12.6 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.8 | 3.9 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.6 | 6.8 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.6 | 3.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.6 | 9.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.6 | 2.2 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.5 | 23.0 | GO:0002102 | podosome(GO:0002102) |
| 0.5 | 3.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.5 | 1.6 | GO:0031251 | PAN complex(GO:0031251) |
| 0.5 | 1.4 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 0.5 | 3.7 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.4 | 9.8 | GO:0043657 | host(GO:0018995) host cell part(GO:0033643) host cell(GO:0043657) |
| 0.4 | 4.8 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.4 | 2.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.4 | 1.8 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.3 | 0.7 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.3 | 2.9 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.3 | 2.9 | GO:0002177 | manchette(GO:0002177) |
| 0.3 | 12.6 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.3 | 1.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.3 | 4.6 | GO:0010369 | chromocenter(GO:0010369) |
| 0.3 | 3.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.3 | 5.5 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.3 | 1.7 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.3 | 5.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.3 | 1.3 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.3 | 0.8 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.3 | 2.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.3 | 3.8 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.3 | 1.8 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.2 | 3.8 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.2 | 1.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.2 | 2.0 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.2 | 1.6 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.2 | 2.0 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.2 | 7.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.2 | 2.5 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.2 | 1.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 1.4 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 0.9 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.2 | 3.0 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 22.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 3.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 0.5 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 0.2 | 5.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 10.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.2 | 12.5 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.2 | 6.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.2 | 0.9 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.2 | 17.3 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 1.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 4.0 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 5.1 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 0.9 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 3.2 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 1.7 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 0.4 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.1 | 1.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 4.6 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 8.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 0.6 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 2.4 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.9 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.8 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 2.0 | GO:0030141 | secretory granule(GO:0030141) |
| 0.1 | 1.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 2.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 4.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 9.5 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 0.6 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.1 | 11.4 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.1 | 0.7 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 1.4 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 0.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.3 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.1 | 2.7 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 1.5 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 4.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 0.7 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 6.6 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.1 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.9 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 0.7 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 1.0 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 2.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 13.8 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 10.5 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 2.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 2.0 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 14.6 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 2.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.6 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 1.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 3.0 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 1.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 1.0 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 2.4 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.7 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 7.4 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 2.2 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.6 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 2.7 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.3 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 1.1 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 1.2 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 1.7 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.6 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 3.5 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 0.3 | GO:0043204 | perikaryon(GO:0043204) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.6 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 3.8 | 11.4 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 2.4 | 21.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 2.0 | 8.2 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 1.8 | 7.0 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 1.6 | 6.3 | GO:1990254 | keratin filament binding(GO:1990254) |
| 1.5 | 16.9 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 1.2 | 14.8 | GO:0031996 | thioesterase binding(GO:0031996) |
| 1.2 | 4.8 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 1.2 | 4.7 | GO:0070976 | calcium-independent protein kinase C activity(GO:0004699) TIR domain binding(GO:0070976) |
| 1.1 | 3.3 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 1.0 | 6.2 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 1.0 | 4.0 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.9 | 2.7 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.9 | 4.5 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.9 | 5.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.8 | 2.5 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.8 | 4.9 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.8 | 3.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.7 | 2.2 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.7 | 7.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.7 | 2.1 | GO:0001565 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.7 | 5.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.7 | 2.7 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.7 | 5.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.7 | 2.7 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.7 | 2.0 | GO:0031779 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.6 | 3.9 | GO:0019841 | retinol binding(GO:0019841) |
| 0.6 | 24.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.6 | 6.7 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.6 | 2.4 | GO:0050632 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.6 | 1.8 | GO:0016901 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.6 | 1.7 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.6 | 12.3 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.6 | 1.7 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.6 | 4.4 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.5 | 2.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.5 | 5.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.5 | 2.6 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.5 | 5.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.5 | 3.9 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.5 | 3.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.5 | 1.4 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.5 | 1.8 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.4 | 18.7 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.4 | 2.6 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.4 | 2.2 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.4 | 3.8 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.4 | 1.9 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.4 | 9.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.4 | 1.5 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.4 | 2.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.4 | 4.3 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.3 | 1.0 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.3 | 2.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.3 | 1.3 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.3 | 5.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.3 | 2.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.3 | 1.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.3 | 2.9 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.3 | 1.3 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.3 | 1.6 | GO:0032356 | oxidized DNA binding(GO:0032356) 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.2 | 3.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 1.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 3.8 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.2 | 0.9 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.2 | 3.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.2 | 1.6 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.2 | 10.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.2 | 4.0 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.2 | 1.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 1.3 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 2.9 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 3.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.2 | 1.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.2 | 1.0 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 1.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.2 | 0.8 | GO:0032405 | MutLalpha complex binding(GO:0032405) |
| 0.2 | 2.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 0.8 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.2 | 21.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 2.5 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.2 | 0.8 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.2 | 3.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 1.6 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.2 | 0.9 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 3.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 1.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.2 | 1.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 0.3 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.2 | 5.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.2 | 1.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 1.8 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.7 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 7.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 1.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 8.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 2.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 1.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.6 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 2.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 5.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 4.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 2.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 5.1 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 3.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 4.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 2.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.4 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 1.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 19.5 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 0.4 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 3.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.6 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.8 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.1 | 1.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 1.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.5 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 0.9 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 1.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 0.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) C-X-C chemokine binding(GO:0019958) |
| 0.1 | 2.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 1.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.2 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 1.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 2.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.2 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 2.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 0.7 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 1.0 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.1 | 1.2 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.1 | 1.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.9 | GO:0052744 | phosphatidylinositol-3-phosphatase activity(GO:0004438) phosphatidylinositol monophosphate phosphatase activity(GO:0052744) |
| 0.1 | 0.2 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 3.6 | GO:0020037 | heme binding(GO:0020037) |
| 0.1 | 30.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 1.0 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.6 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 2.7 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 2.7 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 2.2 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 0.9 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 1.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 36.9 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) |
| 0.1 | 1.6 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.1 | 2.3 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 1.2 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 0.8 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.7 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 2.3 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.4 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.4 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.3 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.1 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.6 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 1.0 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.3 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 1.1 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 1.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 1.4 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 2.4 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 5.1 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 4.6 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.7 | GO:0016597 | amino acid binding(GO:0016597) |
| 0.0 | 2.0 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.2 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 1.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.6 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 13.3 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 0.1 | GO:0017049 | GTP-Rho binding(GO:0017049) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 48.8 | PID_ILK_PATHWAY | Integrin-linked kinase signaling |
| 0.5 | 11.8 | PID_PRL_SIGNALING_EVENTS_PATHWAY | Signaling events mediated by PRL |
| 0.5 | 26.5 | PID_RET_PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.4 | 23.7 | PID_IL12_2PATHWAY | IL12-mediated signaling events |
| 0.4 | 3.9 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.4 | 2.6 | PID_TCR_RAS_PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.4 | 16.5 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.3 | 6.3 | PID_SYNDECAN_2_PATHWAY | Syndecan-2-mediated signaling events |
| 0.3 | 15.0 | PID_PS1_PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.3 | 12.4 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.3 | 1.2 | ST_ADRENERGIC | Adrenergic Pathway |
| 0.3 | 10.6 | ST_G_ALPHA_I_PATHWAY | G alpha i Pathway |
| 0.3 | 8.4 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.3 | 2.8 | PID_ANTHRAX_PATHWAY | Cellular roles of Anthrax toxin |
| 0.2 | 20.6 | WNT_SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.2 | 9.9 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.2 | 2.0 | PID_SYNDECAN_3_PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 4.5 | PID_ALPHA_SYNUCLEIN_PATHWAY | Alpha-synuclein signaling |
| 0.2 | 2.7 | PID_SYNDECAN_4_PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 7.3 | PID_FGF_PATHWAY | FGF signaling pathway |
| 0.2 | 5.0 | PID_FAK_PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.2 | 3.1 | PID_RAC1_PATHWAY | RAC1 signaling pathway |
| 0.1 | 12.5 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 13.4 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 6.8 | PID_CDC42_PATHWAY | CDC42 signaling events |
| 0.1 | 3.2 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.1 | 8.2 | PID_RHOA_REG_PATHWAY | Regulation of RhoA activity |
| 0.1 | 3.4 | PID_AJDISS_2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 7.2 | PID_SMAD2_3NUCLEAR_PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 1.9 | PID_DNA_PK_PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 2.6 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
| 0.1 | 3.3 | PID_AP1_PATHWAY | AP-1 transcription factor network |
| 0.1 | 8.5 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID_CIRCADIAN_PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.3 | PID_THROMBIN_PAR4_PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.0 | PID_EPHB_FWD_PATHWAY | EPHB forward signaling |
| 0.0 | 0.7 | PID_INTEGRIN1_PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 1.0 | PID_VEGFR1_2_PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.2 | PID_REG_GR_PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.6 | PID_RETINOIC_ACID_PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.2 | PID_BCR_5PATHWAY | BCR signaling pathway |
| 0.0 | 0.7 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.2 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.3 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.4 | PID_HDAC_CLASSII_PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.4 | PID_TELOMERASE_PATHWAY | Regulation of Telomerase |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 11.4 | REACTOME_SIGNALING_BY_FGFR3_MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 1.1 | 15.1 | REACTOME_ACTIVATION_OF_IRF3_IRF7_MEDIATED_BY_TBK1_IKK_EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 1.1 | 7.4 | REACTOME_ENDOGENOUS_STEROLS | Genes involved in Endogenous sterols |
| 0.9 | 16.3 | REACTOME_CONVERSION_FROM_APC_C_CDC20_TO_APC_C_CDH1_IN_LATE_ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.7 | 12.1 | REACTOME_NOTCH_HLH_TRANSCRIPTION_PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.6 | 14.0 | REACTOME_CREB_PHOSPHORYLATION_THROUGH_THE_ACTIVATION_OF_CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.5 | 6.8 | REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.4 | 4.5 | REACTOME_AMINE_DERIVED_HORMONES | Genes involved in Amine-derived hormones |
| 0.4 | 5.2 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.4 | 12.1 | REACTOME_G_ALPHA_Z_SIGNALLING_EVENTS | Genes involved in G alpha (z) signalling events |
| 0.4 | 17.5 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.3 | 7.3 | REACTOME_SIGNAL_ATTENUATION | Genes involved in Signal attenuation |
| 0.3 | 10.9 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.3 | 5.3 | REACTOME_TRAF6_MEDIATED_IRF7_ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.3 | 0.3 | REACTOME_ACTIVATED_TAK1_MEDIATES_P38_MAPK_ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.3 | 8.3 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.3 | 3.2 | REACTOME_PRE_NOTCH_TRANSCRIPTION_AND_TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.2 | 6.3 | REACTOME_CELL_EXTRACELLULAR_MATRIX_INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 9.6 | REACTOME_SEMA4D_IN_SEMAPHORIN_SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.2 | 19.8 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.2 | 2.4 | REACTOME_BILE_ACID_AND_BILE_SALT_METABOLISM | Genes involved in Bile acid and bile salt metabolism |
| 0.2 | 4.7 | REACTOME_CASPASE_MEDIATED_CLEAVAGE_OF_CYTOSKELETAL_PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 6.9 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 13.8 | REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.2 | 3.4 | REACTOME_CHONDROITIN_SULFATE_BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.2 | 3.4 | REACTOME_AMINE_COMPOUND_SLC_TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 2.7 | REACTOME_IMMUNOREGULATORY_INTERACTIONS_BETWEEN_A_LYMPHOID_AND_A_NON_LYMPHOID_CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 2.6 | REACTOME_NEGATIVE_REGULATORS_OF_RIG_I_MDA5_SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 2.2 | REACTOME_METABOLISM_OF_POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 4.0 | REACTOME_EXTENSION_OF_TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 1.7 | REACTOME_HDL_MEDIATED_LIPID_TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 2.7 | REACTOME_HS_GAG_DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 1.6 | REACTOME_RESOLUTION_OF_AP_SITES_VIA_THE_SINGLE_NUCLEOTIDE_REPLACEMENT_PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 0.8 | REACTOME_REGULATION_OF_THE_FANCONI_ANEMIA_PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 3.0 | REACTOME_SYNTHESIS_OF_GLYCOSYLPHOSPHATIDYLINOSITOL_GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 2.3 | REACTOME_REGULATION_OF_GENE_EXPRESSION_IN_BETA_CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 5.4 | REACTOME_RNA_POL_I_PROMOTER_OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 2.8 | REACTOME_LATENT_INFECTION_OF_HOMO_SAPIENS_WITH_MYCOBACTERIUM_TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 1.8 | REACTOME_MRNA_DECAY_BY_5_TO_3_EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 0.5 | REACTOME_REGULATION_OF_COMPLEMENT_CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 2.1 | REACTOME_ENOS_ACTIVATION_AND_REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 1.6 | REACTOME_MTORC1_MEDIATED_SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 2.1 | REACTOME_GLUTAMATE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 1.1 | REACTOME_HOMOLOGOUS_RECOMBINATION_REPAIR_OF_REPLICATION_INDEPENDENT_DOUBLE_STRAND_BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 8.0 | REACTOME_G_ALPHA1213_SIGNALLING_EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.1 | 1.5 | REACTOME_INHIBITION_OF_VOLTAGE_GATED_CA2_CHANNELS_VIA_GBETA_GAMMA_SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.4 | REACTOME_ZINC_TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.2 | REACTOME_SIGNALING_BY_ILS | Genes involved in Signaling by Interleukins |
| 0.1 | 1.2 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 0.3 | REACTOME_INHIBITION_OF_REPLICATION_INITIATION_OF_DAMAGED_DNA_BY_RB1_E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 1.4 | REACTOME_FORMATION_OF_INCISION_COMPLEX_IN_GG_NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 1.9 | REACTOME_RNA_POL_III_TRANSCRIPTION_TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 2.1 | REACTOME_SIGNALING_BY_ROBO_RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.7 | REACTOME_CELL_CELL_JUNCTION_ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.1 | 4.8 | REACTOME_G_ALPHA_Q_SIGNALLING_EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 1.7 | REACTOME_NUCLEAR_SIGNALING_BY_ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.4 | REACTOME_PASSIVE_TRANSPORT_BY_AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.8 | REACTOME_REGULATION_OF_GLUCOKINASE_BY_GLUCOKINASE_REGULATORY_PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.3 | REACTOME_INTRINSIC_PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 1.4 | REACTOME_MRNA_3_END_PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 1.0 | REACTOME_RNA_POL_II_TRANSCRIPTION_PRE_INITIATION_AND_PROMOTER_OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.6 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.0 | REACTOME_SYNTHESIS_OF_PA | Genes involved in Synthesis of PA |
| 0.0 | 3.0 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.5 | REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.6 | REACTOME_ENDOSOMAL_SORTING_COMPLEX_REQUIRED_FOR_TRANSPORT_ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 2.0 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME_PYRIMIDINE_METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.1 | REACTOME_JNK_C_JUN_KINASES_PHOSPHORYLATION_AND_ACTIVATION_MEDIATED_BY_ACTIVATED_HUMAN_TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.5 | REACTOME_PYRUVATE_METABOLISM_AND_CITRIC_ACID_TCA_CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |