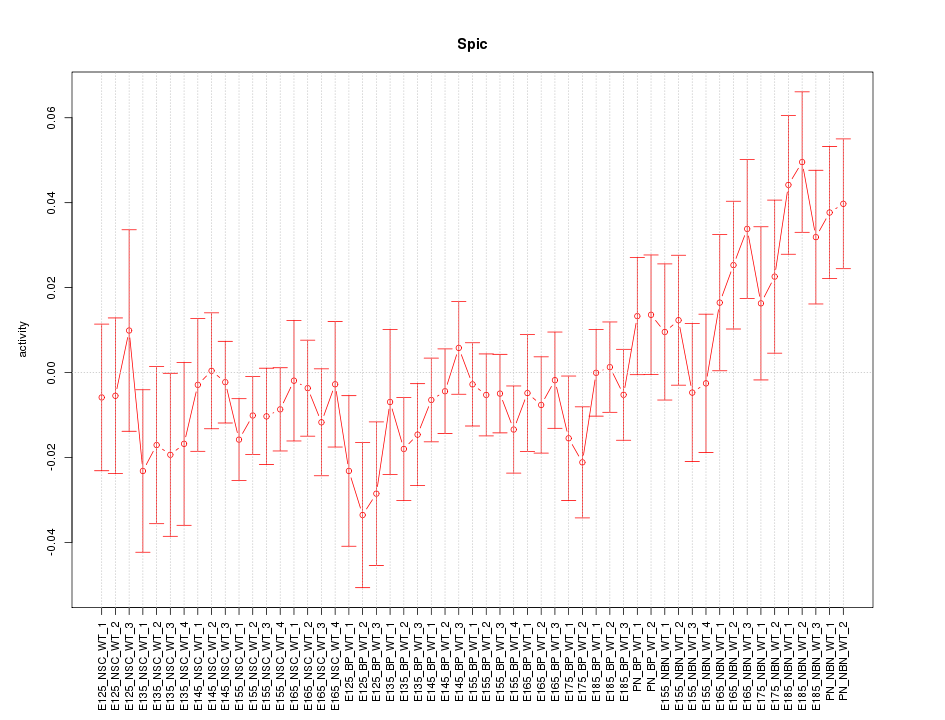
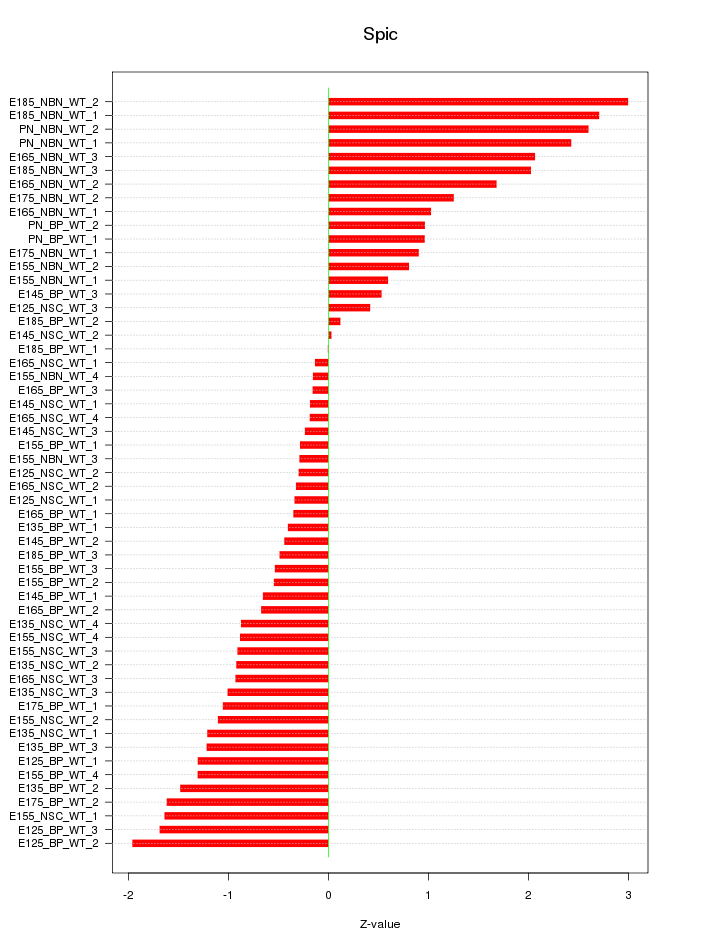
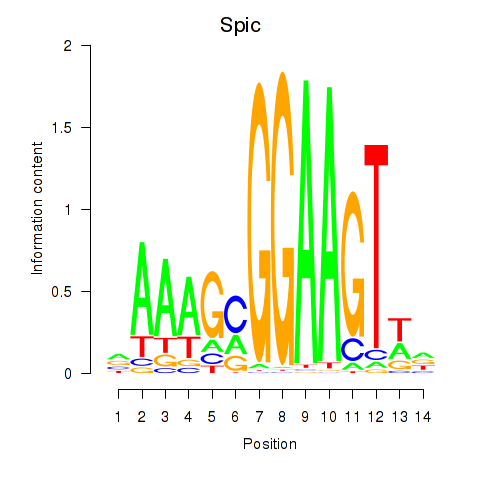
Motif ID: Spic
Z-value: 1.189
Transcription factors associated with Spic:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Spic | ENSMUSG00000004359.10 | Spic |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Spic | mm10_v2_chr10_-_88683021_88683025 | 0.05 | 7.2e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 16.6 | GO:0001803 | type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 2.1 | 15.0 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 1.9 | 9.7 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 1.7 | 5.2 | GO:0016068 | regulation of type I hypersensitivity(GO:0001810) type I hypersensitivity(GO:0016068) |
| 1.6 | 4.8 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 1.6 | 4.7 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 1.5 | 8.9 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 1.4 | 4.2 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 1.3 | 3.9 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 1.3 | 3.9 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 1.0 | 6.1 | GO:0032796 | uropod organization(GO:0032796) |
| 1.0 | 10.7 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 1.0 | 3.8 | GO:0050904 | diapedesis(GO:0050904) |
| 0.9 | 2.6 | GO:2001187 | positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.7 | 2.2 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.7 | 2.2 | GO:2000561 | endocardial cushion fusion(GO:0003274) CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) |
| 0.7 | 2.8 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.7 | 2.8 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.7 | 12.8 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.7 | 9.8 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.6 | 1.8 | GO:2001180 | negative regulation of interleukin-18 production(GO:0032701) negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.6 | 10.0 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.6 | 1.7 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.5 | 3.2 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.5 | 1.5 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.5 | 1.5 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 0.5 | 4.4 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.5 | 1.5 | GO:1902220 | positive regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902220) |
| 0.4 | 0.9 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.4 | 2.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.4 | 1.2 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.4 | 1.2 | GO:1904809 | dense core granule localization(GO:0032253) dense core granule transport(GO:1901950) regulation of dense core granule transport(GO:1904809) positive regulation of dense core granule transport(GO:1904811) |
| 0.4 | 2.0 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.3 | 1.0 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.3 | 3.0 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.3 | 2.3 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.3 | 2.5 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.3 | 0.9 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.3 | 2.4 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.3 | 1.7 | GO:0010730 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.3 | 2.0 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.3 | 4.8 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.3 | 2.8 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.3 | 1.7 | GO:0035878 | nail development(GO:0035878) |
| 0.3 | 0.8 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.3 | 3.5 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.3 | 2.1 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.3 | 1.5 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.3 | 0.8 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.2 | 3.4 | GO:0015816 | glycine transport(GO:0015816) |
| 0.2 | 0.7 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.2 | 2.5 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.2 | 1.8 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.2 | 3.7 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.2 | 0.6 | GO:0045014 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.2 | 0.4 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.2 | 2.2 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.2 | 0.7 | GO:0035826 | rubidium ion transport(GO:0035826) |
| 0.2 | 0.7 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.2 | 2.0 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.2 | 0.7 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.2 | 0.5 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.2 | 2.3 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.2 | 2.7 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.2 | 0.5 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.2 | 0.5 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.2 | 1.7 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.2 | 0.5 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.1 | 1.0 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.1 | 0.3 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.1 | 0.7 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.1 | 3.6 | GO:0000731 | DNA synthesis involved in DNA repair(GO:0000731) |
| 0.1 | 0.6 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 3.7 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.1 | 1.5 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.1 | 3.2 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.1 | 2.5 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 0.7 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 0.4 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.1 | 0.8 | GO:1901911 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 1.5 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 1.3 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.7 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) regulation of peroxisome size(GO:0044375) |
| 0.1 | 2.1 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 4.9 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 7.9 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 0.8 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 0.6 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 3.5 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 0.5 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.9 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.1 | 2.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.5 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.1 | 0.4 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.8 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.1 | 1.8 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 1.0 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 | 0.6 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 1.0 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 0.8 | GO:1990173 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.1 | 0.8 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.1 | 12.3 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.1 | 0.4 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.1 | 0.4 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.1 | 1.8 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 | 0.6 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 0.1 | GO:0039533 | regulation of MDA-5 signaling pathway(GO:0039533) |
| 0.1 | 0.6 | GO:0006544 | glycine metabolic process(GO:0006544) |
| 0.1 | 0.4 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.1 | 0.9 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.1 | 0.8 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 0.3 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.1 | 0.7 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.4 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.1 | 0.4 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 0.4 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.1 | 0.4 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.9 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.1 | 2.0 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 3.4 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.1 | 0.9 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 1.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.7 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.2 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.7 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.3 | GO:0035357 | peroxisome proliferator activated receptor signaling pathway(GO:0035357) |
| 0.0 | 0.9 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.6 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.0 | 0.7 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.0 | 0.7 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.3 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 1.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.5 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.2 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.8 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.2 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 2.0 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.0 | 0.9 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.1 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.7 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.0 | 2.0 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.2 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 | 1.6 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.4 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.6 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.5 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 1.0 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.6 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 1.2 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.8 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 2.8 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 1.0 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 0.0 | GO:1905076 | interleukin-17 secretion(GO:0072615) regulation of interleukin-17 secretion(GO:1905076) |
| 0.0 | 0.7 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.2 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.6 | GO:0006887 | exocytosis(GO:0006887) |
| 0.0 | 0.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.6 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.3 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 0.7 | GO:0002687 | positive regulation of leukocyte migration(GO:0002687) |
| 0.0 | 0.3 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0061470 | interleukin-21 production(GO:0032625) T follicular helper cell differentiation(GO:0061470) interleukin-21 secretion(GO:0072619) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.7 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 1.2 | 3.6 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 1.0 | 3.9 | GO:0044307 | dendritic branch(GO:0044307) |
| 1.0 | 4.8 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.9 | 4.7 | GO:1990745 | EARP complex(GO:1990745) |
| 0.8 | 3.8 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.8 | 4.6 | GO:1990393 | 3M complex(GO:1990393) |
| 0.7 | 3.6 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.7 | 2.1 | GO:0099631 | postsynaptic endocytic zone cytoplasmic component(GO:0099631) |
| 0.6 | 3.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.6 | 6.1 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.5 | 3.0 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.5 | 3.3 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.4 | 1.8 | GO:0098888 | extrinsic component of presynaptic membrane(GO:0098888) |
| 0.4 | 6.2 | GO:0042581 | specific granule(GO:0042581) |
| 0.4 | 1.2 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.4 | 28.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.3 | 2.0 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.3 | 3.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 1.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.2 | 0.7 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.2 | 3.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 2.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.2 | 7.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 0.6 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.2 | 1.4 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.2 | 1.5 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.2 | 2.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.2 | 4.6 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 0.9 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.2 | 0.5 | GO:0055087 | Ski complex(GO:0055087) |
| 0.2 | 1.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.2 | 1.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.2 | 1.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 1.0 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 0.6 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.1 | 2.0 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 2.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.8 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.9 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 3.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 25.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 7.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.3 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.1 | 1.0 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 2.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.8 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 0.6 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 0.4 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 7.6 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 0.8 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.7 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 2.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 7.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 0.1 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.1 | 13.3 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 0.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.4 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 0.6 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 0.3 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 1.6 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.2 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.0 | 2.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.8 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.1 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 1.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 2.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 2.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.5 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 6.5 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.9 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.8 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0048500 | signal recognition particle(GO:0048500) |
| 0.0 | 0.8 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 1.0 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.3 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 1.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.5 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 2.5 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.7 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.8 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.2 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.2 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.7 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.7 | GO:0045177 | apical part of cell(GO:0045177) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 21.8 | GO:0019864 | immunoglobulin receptor activity(GO:0019763) IgG binding(GO:0019864) |
| 1.6 | 4.8 | GO:0003796 | lysozyme activity(GO:0003796) |
| 1.4 | 4.2 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 1.3 | 8.9 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 1.1 | 3.2 | GO:0016901 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 1.0 | 6.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.9 | 3.6 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.7 | 7.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.7 | 2.8 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.6 | 10.7 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.6 | 2.4 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.6 | 2.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.6 | 6.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.5 | 3.6 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.5 | 3.0 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.5 | 2.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.4 | 2.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.4 | 2.5 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.4 | 3.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.4 | 1.2 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.4 | 1.2 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.3 | 2.8 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.3 | 3.4 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.3 | 0.9 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.3 | 1.8 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.3 | 1.2 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.3 | 0.9 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.3 | 3.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.3 | 1.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.3 | 9.2 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.2 | 7.9 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.2 | 0.7 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.2 | 1.3 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.2 | 0.7 | GO:0004534 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) 5'-3' exoribonuclease activity(GO:0004534) |
| 0.2 | 2.9 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 0.8 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.2 | 7.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.2 | 4.1 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.2 | 1.8 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.2 | 3.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.2 | 2.1 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.2 | 0.5 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.2 | 3.9 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.2 | 0.7 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.2 | 4.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.7 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 1.5 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 1.3 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 4.6 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 0.8 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 1.5 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.1 | 0.4 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 0.8 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 1.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.9 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 0.6 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.9 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 0.4 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.1 | 0.7 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 1.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 4.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 0.4 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 1.7 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.6 | GO:0005047 | signal recognition particle binding(GO:0005047) 7S RNA binding(GO:0008312) |
| 0.1 | 2.2 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.1 | 0.9 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.1 | 8.6 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 1.0 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.4 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.1 | 2.3 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 1.8 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 0.8 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 0.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 2.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 1.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 3.7 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 0.7 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 2.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.8 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 1.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.2 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.6 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 1.0 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.9 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.1 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.0 | 2.8 | GO:0019209 | kinase activator activity(GO:0019209) |
| 0.0 | 1.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.8 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.0 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 2.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.6 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 1.0 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.4 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 3.8 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.8 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 1.0 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.9 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.2 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.5 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 2.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 5.9 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.8 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.9 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.5 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.3 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 1.0 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.0 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.2 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 1.8 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.1 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.6 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 8.3 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.2 | GO:0030276 | clathrin binding(GO:0030276) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.3 | PID_TCR_JNK_PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.3 | 34.0 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.3 | 22.6 | PID_FCER1_PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.2 | 9.3 | PID_KIT_PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 2.5 | PID_LPA4_PATHWAY | LPA4-mediated signaling events |
| 0.1 | 7.6 | PID_RAC1_PATHWAY | RAC1 signaling pathway |
| 0.1 | 6.5 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.1 | 3.5 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.1 | 0.6 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 6.7 | PID_RHOA_REG_PATHWAY | Regulation of RhoA activity |
| 0.1 | 2.3 | ST_ADRENERGIC | Adrenergic Pathway |
| 0.1 | 0.8 | PID_VEGF_VEGFR_PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 3.6 | PID_INTEGRIN3_PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 7.3 | PID_PDGFRB_PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 2.0 | PID_DNA_PK_PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 4.6 | PID_HDAC_CLASSII_PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 0.5 | PID_EPO_PATHWAY | EPO signaling pathway |
| 0.0 | 0.3 | PID_ANTHRAX_PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 6.5 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.7 | PID_ERB_GENOMIC_PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.2 | SA_B_CELL_RECEPTOR_COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 5.9 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.0 | PID_VEGFR1_2_PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.7 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.4 | PID_P38_GAMMA_DELTA_PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.6 | WNT_SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.6 | PID_CDC42_PATHWAY | CDC42 signaling events |
| 0.0 | 0.6 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.9 | PID_MYC_REPRESS_PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.6 | PID_AVB3_INTEGRIN_PATHWAY | Integrins in angiogenesis |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 29.0 | REACTOME_CREATION_OF_C4_AND_C2_ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 2.9 | 8.7 | REACTOME_ENDOSOMAL_VACUOLAR_PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.8 | 10.4 | REACTOME_PLATELET_ADHESION_TO_EXPOSED_COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.4 | 3.6 | REACTOME_P2Y_RECEPTORS | Genes involved in P2Y receptors |
| 0.3 | 3.0 | REACTOME_REMOVAL_OF_THE_FLAP_INTERMEDIATE_FROM_THE_C_STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.3 | 5.2 | REACTOME_IMMUNOREGULATORY_INTERACTIONS_BETWEEN_A_LYMPHOID_AND_A_NON_LYMPHOID_CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.2 | 6.8 | REACTOME_LATENT_INFECTION_OF_HOMO_SAPIENS_WITH_MYCOBACTERIUM_TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.2 | 3.8 | REACTOME_PLATELET_SENSITIZATION_BY_LDL | Genes involved in Platelet sensitization by LDL |
| 0.2 | 2.5 | REACTOME_ADENYLATE_CYCLASE_ACTIVATING_PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.2 | 1.5 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 1.5 | REACTOME_TRAFFICKING_AND_PROCESSING_OF_ENDOSOMAL_TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.2 | 3.0 | REACTOME_INTEGRATION_OF_PROVIRUS | Genes involved in Integration of provirus |
| 0.2 | 4.1 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION_IN_THE_MEDIAL_TRANS_GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.2 | 3.9 | REACTOME_CREB_PHOSPHORYLATION_THROUGH_THE_ACTIVATION_OF_CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 1.9 | REACTOME_PROTEOLYTIC_CLEAVAGE_OF_SNARE_COMPLEX_PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.5 | REACTOME_REGULATION_OF_RHEB_GTPASE_ACTIVITY_BY_AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 2.1 | REACTOME_GAP_JUNCTION_DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 0.8 | REACTOME_FGFR4_LIGAND_BINDING_AND_ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 3.4 | REACTOME_AMINO_ACID_TRANSPORT_ACROSS_THE_PLASMA_MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 4.0 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 2.2 | REACTOME_TGF_BETA_RECEPTOR_SIGNALING_IN_EMT_EPITHELIAL_TO_MESENCHYMAL_TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 3.0 | REACTOME_SYNTHESIS_OF_PA | Genes involved in Synthesis of PA |
| 0.1 | 15.3 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.2 | REACTOME_ADVANCED_GLYCOSYLATION_ENDPRODUCT_RECEPTOR_SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 0.4 | REACTOME_PROCESSIVE_SYNTHESIS_ON_THE_LAGGING_STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.1 | 3.3 | REACTOME_INWARDLY_RECTIFYING_K_CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 0.7 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PC | Genes involved in Acyl chain remodelling of PC |
| 0.1 | 0.8 | REACTOME_VEGF_LIGAND_RECEPTOR_INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 2.8 | REACTOME_TRANSPORT_OF_VITAMINS_NUCLEOSIDES_AND_RELATED_MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 1.8 | REACTOME_EGFR_DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 5.5 | REACTOME_MHC_CLASS_II_ANTIGEN_PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 0.7 | REACTOME_P130CAS_LINKAGE_TO_MAPK_SIGNALING_FOR_INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 0.9 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_GOLGI_MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 4.4 | REACTOME_PEPTIDE_CHAIN_ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.4 | REACTOME_REGULATION_OF_WATER_BALANCE_BY_RENAL_AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 1.0 | REACTOME_PHASE1_FUNCTIONALIZATION_OF_COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.4 | REACTOME_COPI_MEDIATED_TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.4 | REACTOME_BILE_ACID_AND_BILE_SALT_METABOLISM | Genes involved in Bile acid and bile salt metabolism |
| 0.0 | 0.6 | REACTOME_FORMATION_OF_INCISION_COMPLEX_IN_GG_NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.0 | REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_GLUCAGON_LIKE_PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.5 | REACTOME_ANTIGEN_ACTIVATES_B_CELL_RECEPTOR_LEADING_TO_GENERATION_OF_SECOND_MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.7 | REACTOME_SYNTHESIS_OF_VERY_LONG_CHAIN_FATTY_ACYL_COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.3 | REACTOME_GLYCEROPHOSPHOLIPID_BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 3.5 | REACTOME_FATTY_ACID_TRIACYLGLYCEROL_AND_KETONE_BODY_METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 1.8 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.6 | REACTOME_REGULATION_OF_GENE_EXPRESSION_IN_BETA_CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 2.1 | REACTOME_RNA_POL_I_RNA_POL_III_AND_MITOCHONDRIAL_TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 0.3 | REACTOME_PURINE_SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.7 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.2 | REACTOME_IRON_UPTAKE_AND_TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.5 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |