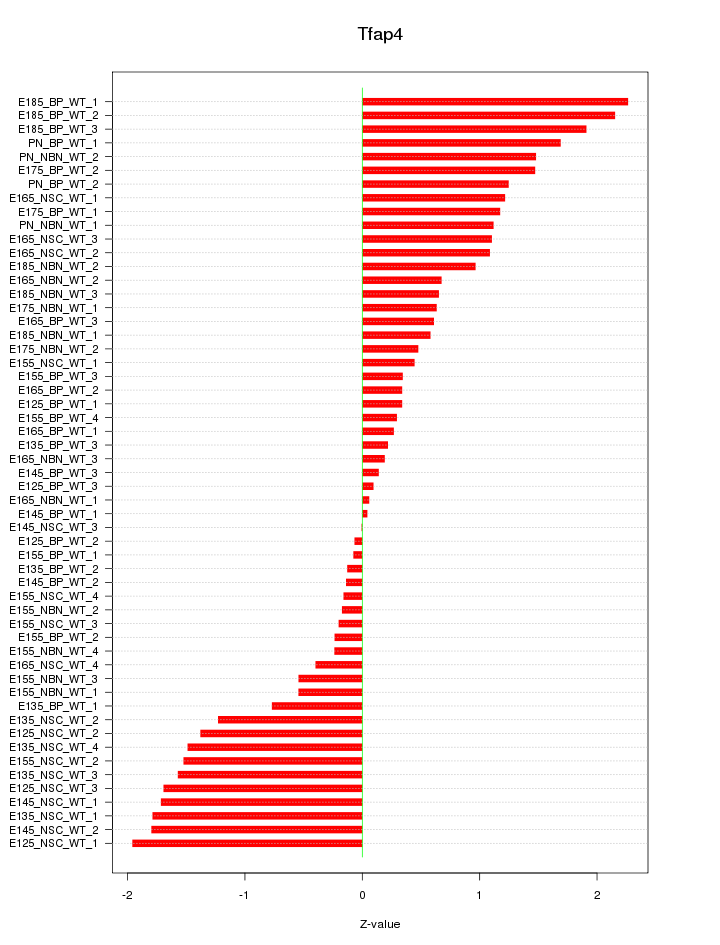
Motif ID: Tfap4
Z-value: 1.051
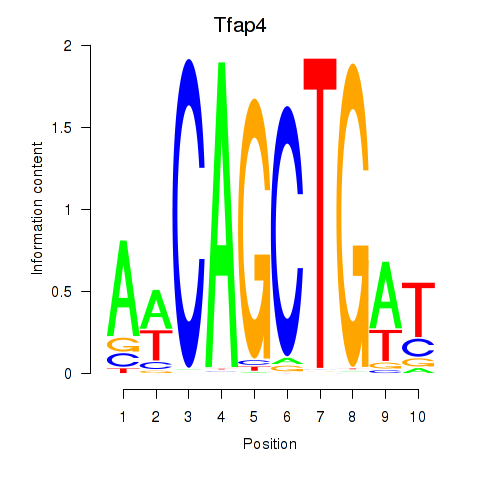
Transcription factors associated with Tfap4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Tfap4 | ENSMUSG00000005718.7 | Tfap4 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfap4 | mm10_v2_chr16_-_4559720_4559747 | -0.16 | 2.3e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.9 | GO:0038095 | positive regulation of mast cell cytokine production(GO:0032765) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 2.0 | 8.0 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 1.8 | 9.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 1.8 | 7.0 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.8 | 2.5 | GO:0003104 | positive regulation of glomerular filtration(GO:0003104) negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) phenotypic switching(GO:0036166) metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 0.8 | 4.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) negative regulation of cytolysis(GO:0045918) |
| 0.8 | 2.3 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.8 | 2.3 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.7 | 2.2 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.7 | 2.8 | GO:1904451 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.6 | 1.9 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.6 | 1.9 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.5 | 3.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.5 | 1.4 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 0.5 | 8.1 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.5 | 3.7 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.4 | 3.0 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.4 | 3.8 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.4 | 2.2 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.4 | 1.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.4 | 3.3 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.3 | 1.0 | GO:0071336 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.3 | 5.5 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.3 | 1.7 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.3 | 3.6 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.3 | 1.0 | GO:0046543 | development of secondary sexual characteristics(GO:0045136) development of secondary female sexual characteristics(GO:0046543) |
| 0.3 | 0.9 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.3 | 0.9 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.3 | 2.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.3 | 1.2 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.3 | 1.8 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.3 | 2.0 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.3 | 2.5 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.3 | 0.8 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) |
| 0.3 | 1.4 | GO:1902995 | apolipoprotein A-I-mediated signaling pathway(GO:0038027) regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.3 | 1.6 | GO:1903056 | positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.3 | 0.8 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.3 | 1.3 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.2 | 1.0 | GO:0046709 | IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.2 | 1.7 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.2 | 0.7 | GO:1904046 | seryl-tRNA aminoacylation(GO:0006434) negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.2 | 1.2 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.2 | 1.2 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.2 | 0.2 | GO:0080144 | amino acid homeostasis(GO:0080144) |
| 0.2 | 3.0 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.2 | 0.6 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.2 | 0.6 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.2 | 2.4 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.2 | 0.6 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.2 | 0.6 | GO:1904760 | myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.2 | 1.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.2 | 0.9 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.2 | 5.6 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.2 | 6.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 0.7 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.2 | 1.1 | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.2 | 4.7 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.2 | 4.4 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.2 | 1.3 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.2 | 2.0 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.2 | 4.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 1.5 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.1 | 1.2 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 2.1 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.1 | 2.8 | GO:0021681 | cerebellar granular layer development(GO:0021681) |
| 0.1 | 1.6 | GO:0060742 | prostate gland growth(GO:0060736) epithelial cell differentiation involved in prostate gland development(GO:0060742) |
| 0.1 | 3.2 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 | 3.5 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 0.8 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 2.4 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.1 | 1.9 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.1 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.1 | 6.5 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 1.7 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 1.0 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 7.7 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 4.7 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.1 | 1.2 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 | 0.5 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 1.1 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 0.8 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 1.5 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 4.0 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 2.1 | GO:0071357 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 | 0.4 | GO:0010155 | regulation of proton transport(GO:0010155) |
| 0.1 | 2.2 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.1 | 1.1 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.1 | 0.7 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 0.7 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.4 | GO:1990034 | regulation of skeletal muscle contraction(GO:0014819) calcium ion export from cell(GO:1990034) |
| 0.1 | 4.5 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 2.4 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 1.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 2.4 | GO:0021799 | cerebral cortex radially oriented cell migration(GO:0021799) centrosome localization(GO:0051642) |
| 0.1 | 1.1 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.1 | 0.8 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.1 | 1.8 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.1 | 0.8 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.1 | 4.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 1.4 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.2 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.1 | 1.0 | GO:0033006 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.1 | 1.8 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 0.4 | GO:1900194 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 2.1 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.1 | 5.1 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.1 | 0.3 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 | 0.7 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 1.9 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 0.3 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 | 2.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.4 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 2.8 | GO:0030518 | intracellular steroid hormone receptor signaling pathway(GO:0030518) |
| 0.0 | 0.3 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.5 | GO:0014898 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 0.8 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 2.1 | GO:0055072 | iron ion homeostasis(GO:0055072) |
| 0.0 | 1.4 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.7 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 1.3 | GO:1901264 | carbohydrate derivative transport(GO:1901264) |
| 0.0 | 1.4 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 1.0 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 1.8 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.0 | 2.0 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 3.2 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 2.7 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 2.1 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.1 | GO:0010575 | positive regulation of vascular endothelial growth factor production(GO:0010575) |
| 0.0 | 0.1 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.0 | 0.5 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.0 | 1.5 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 1.1 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.5 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.5 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:0097574 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) |
| 0.6 | 2.2 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 0.5 | 3.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.5 | 2.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.5 | 7.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.4 | 3.8 | GO:0005883 | neurofilament(GO:0005883) |
| 0.4 | 4.3 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.3 | 1.7 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.3 | 0.6 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.3 | 1.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.3 | 1.2 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.3 | 1.3 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.3 | 8.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 1.7 | GO:0071203 | WASH complex(GO:0071203) |
| 0.2 | 3.8 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.2 | 1.9 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.2 | 0.7 | GO:0098830 | presynaptic endosome(GO:0098830) |
| 0.2 | 3.0 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.2 | 3.8 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 0.8 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 1.5 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.2 | 1.5 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.2 | 3.6 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.2 | 1.4 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.2 | 0.7 | GO:0002141 | stereocilia ankle link(GO:0002141) |
| 0.2 | 0.8 | GO:0008091 | spectrin(GO:0008091) cuticular plate(GO:0032437) |
| 0.2 | 3.0 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.2 | 1.7 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.8 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 1.6 | GO:0031258 | catenin complex(GO:0016342) lamellipodium membrane(GO:0031258) |
| 0.1 | 0.8 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 4.6 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 0.8 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 4.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 3.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 11.1 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 1.2 | GO:0044754 | secondary lysosome(GO:0005767) autolysosome(GO:0044754) |
| 0.1 | 1.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 12.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 1.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 1.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.5 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 1.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.9 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 1.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 7.4 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 1.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 6.8 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 2.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.4 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 1.7 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.3 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.1 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 0.0 | 0.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 2.7 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.8 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 1.7 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.0 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 1.8 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.7 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 3.5 | GO:0030425 | dendrite(GO:0030425) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.7 | GO:0004952 | dopamine neurotransmitter receptor activity(GO:0004952) |
| 1.1 | 4.4 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 1.0 | 9.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 1.0 | 3.0 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.9 | 8.0 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.9 | 7.0 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.8 | 4.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.8 | 2.4 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.6 | 3.7 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.5 | 2.0 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.5 | 2.3 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.5 | 1.4 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.4 | 1.8 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.4 | 2.5 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.4 | 2.0 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.4 | 1.9 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.4 | 1.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.4 | 1.5 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.4 | 3.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.3 | 2.8 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.3 | 3.0 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.3 | 8.1 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.3 | 1.6 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.3 | 1.3 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.3 | 2.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.3 | 1.9 | GO:0009374 | biotin binding(GO:0009374) |
| 0.3 | 6.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.3 | 1.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.3 | 4.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.3 | 1.0 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.2 | 4.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 0.7 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.2 | 1.2 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.2 | 1.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 0.9 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.2 | 1.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.2 | 1.7 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.2 | 6.5 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.2 | 6.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.2 | 2.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 6.9 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.2 | 1.0 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.2 | 3.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 1.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.8 | GO:0048038 | quinone binding(GO:0048038) |
| 0.1 | 0.6 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 2.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 4.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 4.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 1.9 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 2.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.3 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.1 | 1.0 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.1 | 0.8 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.1 | 0.4 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.7 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 1.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 3.8 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 2.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 0.8 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 1.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 4.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.7 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.1 | 0.3 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 2.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.2 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.4 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.0 | 1.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.6 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.6 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 3.8 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 1.3 | GO:1901505 | carbohydrate derivative transporter activity(GO:1901505) |
| 0.0 | 0.9 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 2.7 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 1.2 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 1.4 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 1.2 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 1.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.5 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.2 | GO:0031702 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 1.1 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0061733 | peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 1.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.2 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | ST_TYPE_I_INTERFERON_PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.2 | 6.9 | ST_GA12_PATHWAY | G alpha 12 Pathway |
| 0.2 | 2.2 | PID_VEGF_VEGFR_PATHWAY | VEGF and VEGFR signaling network |
| 0.2 | 2.8 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.2 | 9.0 | PID_REELIN_PATHWAY | Reelin signaling pathway |
| 0.1 | 5.5 | PID_RET_PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 2.5 | PID_S1P_S1P1_PATHWAY | S1P1 pathway |
| 0.1 | 4.1 | PID_CXCR3_PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 6.3 | PID_TRKR_PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 2.8 | PID_FAS_PATHWAY | FAS (CD95) signaling pathway |
| 0.1 | 5.4 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 1.3 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 3.2 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 3.7 | PID_AP1_PATHWAY | AP-1 transcription factor network |
| 0.0 | 2.2 | PID_MYC_REPRESS_PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.6 | PID_GLYPICAN_1PATHWAY | Glypican 1 network |
| 0.0 | 0.7 | PID_IL1_PATHWAY | IL1-mediated signaling events |
| 0.0 | 5.9 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.5 | PID_ERBB2_ERBB3_PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 3.7 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.6 | PID_PDGFRB_PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.6 | PID_KIT_PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 2.0 | WNT_SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.5 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.0 | 2.4 | NABA_MATRISOME_ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 0.9 | PID_SMAD2_3NUCLEAR_PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | PID_HIF1A_PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.1 | REACTOME_COMMON_PATHWAY | Genes involved in Common Pathway |
| 0.3 | 2.3 | REACTOME_GAMMA_CARBOXYLATION_TRANSPORT_AND_AMINO_TERMINAL_CLEAVAGE_OF_PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.3 | 3.7 | REACTOME_ROLE_OF_DCC_IN_REGULATING_APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.3 | 7.0 | REACTOME_CREB_PHOSPHORYLATION_THROUGH_THE_ACTIVATION_OF_CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.2 | 2.2 | REACTOME_VEGF_LIGAND_RECEPTOR_INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 2.1 | REACTOME_REGULATION_OF_IFNA_SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 1.7 | REACTOME_PROTEOLYTIC_CLEAVAGE_OF_SNARE_COMPLEX_PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 4.4 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.9 | REACTOME_SYNTHESIS_SECRETION_AND_DEACYLATION_OF_GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 0.8 | REACTOME_THE_ACTIVATION_OF_ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 2.4 | REACTOME_TRANSFERRIN_ENDOCYTOSIS_AND_RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.1 | 4.4 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.8 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 2.4 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.2 | REACTOME_PLATELET_ADHESION_TO_EXPOSED_COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 0.6 | REACTOME_THE_NLRP3_INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.0 | REACTOME_FGFR1_LIGAND_BINDING_AND_ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 2.6 | REACTOME_ABC_FAMILY_PROTEINS_MEDIATED_TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 1.1 | REACTOME_REGULATION_OF_PYRUVATE_DEHYDROGENASE_PDH_COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 2.8 | REACTOME_RAP1_SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 1.9 | REACTOME_SYNTHESIS_OF_PC | Genes involved in Synthesis of PC |
| 0.1 | 2.0 | REACTOME_AMINO_ACID_SYNTHESIS_AND_INTERCONVERSION_TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 5.8 | REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 2.3 | REACTOME_SYNTHESIS_OF_PA | Genes involved in Synthesis of PA |
| 0.1 | 0.5 | REACTOME_PRESYNAPTIC_NICOTINIC_ACETYLCHOLINE_RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.4 | REACTOME_FRS2_MEDIATED_CASCADE | Genes involved in FRS2-mediated cascade |
| 0.1 | 1.6 | REACTOME_GLYCOSPHINGOLIPID_METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.4 | REACTOME_RNA_POL_III_TRANSCRIPTION_INITIATION_FROM_TYPE_2_PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 1.1 | REACTOME_BIOSYNTHESIS_OF_THE_N_GLYCAN_PRECURSOR_DOLICHOL_LIPID_LINKED_OLIGOSACCHARIDE_LLO_AND_TRANSFER_TO_A_NASCENT_PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.7 | REACTOME_REGULATION_OF_SIGNALING_BY_CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.4 | REACTOME_PLATELET_CALCIUM_HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 1.2 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.2 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.7 | REACTOME_TRANSPORT_TO_THE_GOLGI_AND_SUBSEQUENT_MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.7 | REACTOME_CYTOSOLIC_TRNA_AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.8 | REACTOME_SEMA4D_INDUCED_CELL_MIGRATION_AND_GROWTH_CONE_COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.3 | REACTOME_AMINO_ACID_AND_OLIGOPEPTIDE_SLC_TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.7 | REACTOME_IL1_SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.2 | REACTOME_E2F_ENABLED_INHIBITION_OF_PRE_REPLICATION_COMPLEX_FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.2 | REACTOME_GABA_B_RECEPTOR_ACTIVATION | Genes involved in GABA B receptor activation |
| 0.0 | 0.4 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.8 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |