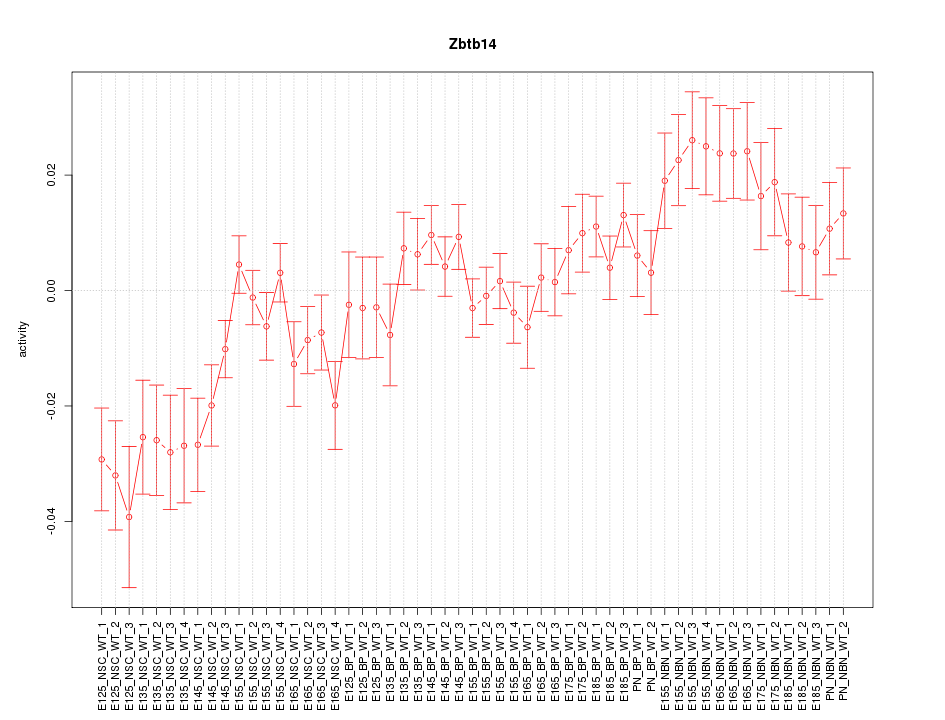
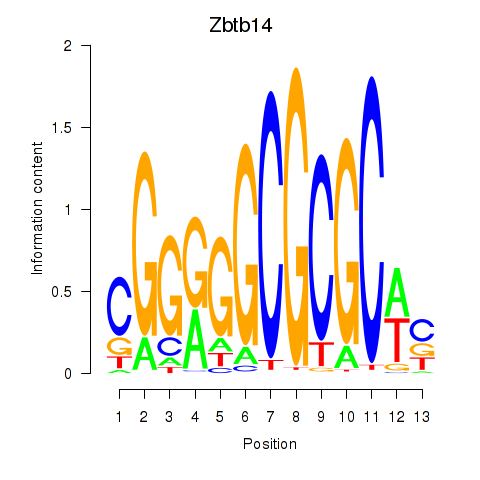
Motif ID: Zbtb14
Z-value: 1.906
Transcription factors associated with Zbtb14:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Zbtb14 | ENSMUSG00000049672.8 | Zbtb14 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb14 | mm10_v2_chr17_+_69383024_69383065 | 0.25 | 6.7e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.4 | 28.3 | GO:0032430 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) positive regulation of phospholipase A2 activity(GO:0032430) activation of meiosis involved in egg activation(GO:0060466) |
| 6.2 | 56.0 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 6.1 | 18.2 | GO:1902512 | positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 5.4 | 32.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 5.3 | 15.8 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) |
| 4.1 | 4.1 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 3.6 | 14.4 | GO:0021564 | glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 3.3 | 9.9 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 3.3 | 9.8 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 3.1 | 9.4 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 3.1 | 3.1 | GO:0002931 | response to ischemia(GO:0002931) |
| 3.1 | 9.3 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 3.0 | 9.1 | GO:1901608 | regulation of vesicle transport along microtubule(GO:1901608) |
| 2.9 | 5.8 | GO:0086047 | membrane depolarization during Purkinje myocyte cell action potential(GO:0086047) |
| 2.9 | 8.6 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 2.8 | 8.4 | GO:0038095 | positive regulation of mast cell cytokine production(GO:0032765) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 2.5 | 10.1 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 2.5 | 5.0 | GO:0035624 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 2.4 | 7.3 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 2.4 | 7.1 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 2.3 | 7.0 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 2.3 | 6.8 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 2.2 | 8.8 | GO:0002924 | negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) |
| 2.0 | 7.9 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 2.0 | 5.9 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 1.9 | 7.8 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 1.9 | 5.7 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 1.8 | 7.3 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 1.7 | 10.4 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 1.7 | 5.0 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 1.6 | 4.9 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 1.6 | 4.8 | GO:0030070 | insulin processing(GO:0030070) |
| 1.5 | 6.1 | GO:0044805 | late nucleophagy(GO:0044805) |
| 1.5 | 10.5 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 1.5 | 2.9 | GO:0002159 | desmosome assembly(GO:0002159) |
| 1.5 | 11.7 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 1.4 | 1.4 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 1.4 | 4.2 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 1.4 | 7.0 | GO:0071692 | protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 1.4 | 4.2 | GO:0016598 | protein arginylation(GO:0016598) |
| 1.4 | 4.1 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 1.4 | 4.1 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 1.4 | 2.7 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 1.3 | 2.7 | GO:0032650 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) |
| 1.3 | 5.4 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 1.3 | 5.2 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 1.3 | 2.6 | GO:1902287 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 1.2 | 3.7 | GO:1990927 | calcium ion regulated lysosome exocytosis(GO:1990927) |
| 1.2 | 14.8 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 1.2 | 8.6 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 1.2 | 7.3 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 1.2 | 15.8 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 1.2 | 3.6 | GO:2000331 | regulation of terminal button organization(GO:2000331) |
| 1.2 | 13.2 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 1.2 | 3.5 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) |
| 1.2 | 12.7 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 1.1 | 4.5 | GO:0097298 | regulation of nucleus size(GO:0097298) |
| 1.1 | 3.2 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 1.1 | 17.1 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 1.0 | 3.1 | GO:0061744 | motor behavior(GO:0061744) |
| 1.0 | 4.1 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 1.0 | 3.1 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) regulation of connective tissue replacement(GO:1905203) |
| 1.0 | 2.9 | GO:1900453 | negative regulation of long term synaptic depression(GO:1900453) |
| 0.9 | 3.8 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.9 | 3.7 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.9 | 4.5 | GO:2000551 | regulation of T-helper 2 cell cytokine production(GO:2000551) positive regulation of T-helper 2 cell cytokine production(GO:2000553) |
| 0.9 | 23.1 | GO:0030204 | chondroitin sulfate metabolic process(GO:0030204) |
| 0.9 | 2.7 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.9 | 2.6 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.9 | 5.3 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.9 | 4.4 | GO:1905247 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.9 | 1.7 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.9 | 4.3 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.9 | 6.0 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.9 | 3.4 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.8 | 3.4 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.8 | 7.6 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.8 | 7.3 | GO:0014010 | Schwann cell proliferation(GO:0014010) |
| 0.8 | 4.0 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.8 | 3.2 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.8 | 2.4 | GO:0035844 | positive regulation of polarized epithelial cell differentiation(GO:0030862) cloaca development(GO:0035844) |
| 0.8 | 3.2 | GO:1900220 | negative regulation of alkaline phosphatase activity(GO:0010693) semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.8 | 2.3 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.8 | 2.3 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.8 | 2.3 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 0.8 | 0.8 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.8 | 5.3 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.7 | 3.7 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.7 | 2.2 | GO:0035864 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.7 | 3.6 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.7 | 7.2 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.7 | 1.4 | GO:0061357 | Wnt protein secretion(GO:0061355) regulation of Wnt protein secretion(GO:0061356) positive regulation of Wnt protein secretion(GO:0061357) |
| 0.7 | 1.4 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.7 | 1.4 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.7 | 19.8 | GO:0095500 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.7 | 7.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.7 | 2.1 | GO:0042748 | circadian sleep/wake cycle, non-REM sleep(GO:0042748) |
| 0.7 | 2.8 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.7 | 2.1 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.7 | 12.9 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.7 | 6.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.7 | 4.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.7 | 12.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.7 | 7.3 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.7 | 4.0 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.7 | 4.6 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.7 | 3.9 | GO:0075044 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.7 | 3.9 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.6 | 1.3 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.6 | 18.8 | GO:0061098 | positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.6 | 5.8 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.6 | 1.3 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 0.6 | 1.9 | GO:0072720 | cellular response to UV-A(GO:0071492) response to dithiothreitol(GO:0072720) |
| 0.6 | 1.2 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.6 | 6.1 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.6 | 1.8 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.6 | 2.4 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.6 | 3.7 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.6 | 2.4 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.6 | 8.4 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.6 | 5.4 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.6 | 1.7 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.6 | 8.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.6 | 7.5 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.6 | 2.2 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.5 | 3.3 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.5 | 2.2 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.5 | 1.6 | GO:0048865 | stem cell fate commitment(GO:0048865) stem cell fate specification(GO:0048866) |
| 0.5 | 2.7 | GO:0032423 | regulation of mismatch repair(GO:0032423) |
| 0.5 | 2.6 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.5 | 14.6 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.5 | 4.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.5 | 2.5 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.5 | 4.5 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.5 | 3.5 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.5 | 2.0 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.5 | 1.0 | GO:2000850 | negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.5 | 3.9 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.5 | 6.2 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.5 | 3.3 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.5 | 1.4 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.5 | 2.8 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.5 | 6.0 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.5 | 4.2 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.5 | 1.8 | GO:1904528 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) positive regulation of microtubule binding(GO:1904528) |
| 0.5 | 1.4 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.5 | 3.7 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.5 | 1.4 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.4 | 5.4 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.4 | 5.2 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.4 | 5.2 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.4 | 5.2 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.4 | 1.7 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.4 | 1.7 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.4 | 2.5 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.4 | 2.5 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.4 | 2.5 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.4 | 8.9 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.4 | 1.3 | GO:0030300 | regulation of intestinal cholesterol absorption(GO:0030300) |
| 0.4 | 0.4 | GO:0003256 | regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003256) |
| 0.4 | 3.4 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.4 | 10.0 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.4 | 2.5 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.4 | 2.9 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.4 | 1.2 | GO:0006507 | GPI anchor release(GO:0006507) |
| 0.4 | 16.8 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.4 | 1.6 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.4 | 1.2 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.4 | 1.2 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.4 | 1.6 | GO:1903859 | regulation of dendrite extension(GO:1903859) |
| 0.4 | 1.2 | GO:0009182 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) GDP metabolic process(GO:0046710) |
| 0.4 | 1.9 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.4 | 7.7 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.4 | 4.9 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.4 | 0.8 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.4 | 1.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.4 | 1.5 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.4 | 4.8 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.4 | 5.1 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.4 | 6.1 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.4 | 5.0 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.4 | 2.5 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.4 | 0.7 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.4 | 1.4 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.3 | 1.7 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.3 | 1.7 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.3 | 2.8 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.3 | 2.8 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.3 | 3.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.3 | 1.4 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.3 | 1.0 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.3 | 1.0 | GO:0071455 | cellular response to hyperoxia(GO:0071455) |
| 0.3 | 8.0 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 4.6 | GO:0001964 | startle response(GO:0001964) |
| 0.3 | 1.6 | GO:2000858 | mineralocorticoid secretion(GO:0035931) aldosterone secretion(GO:0035932) regulation of mineralocorticoid secretion(GO:2000855) positive regulation of mineralocorticoid secretion(GO:2000857) regulation of aldosterone secretion(GO:2000858) positive regulation of aldosterone secretion(GO:2000860) |
| 0.3 | 1.3 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.3 | 0.3 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.3 | 3.4 | GO:0043691 | reverse cholesterol transport(GO:0043691) |
| 0.3 | 1.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.3 | 0.9 | GO:1901166 | calcium-independent cell-matrix adhesion(GO:0007161) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.3 | 1.2 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.3 | 1.2 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.3 | 5.3 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.3 | 3.3 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.3 | 1.8 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.3 | 1.2 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.3 | 1.5 | GO:1902510 | regulation of apoptotic DNA fragmentation(GO:1902510) |
| 0.3 | 2.0 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.3 | 5.2 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.3 | 0.9 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.3 | 3.4 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.3 | 1.1 | GO:0010896 | negative regulation of sequestering of triglyceride(GO:0010891) regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.3 | 0.6 | GO:0090204 | protein localization to nuclear pore(GO:0090204) positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.3 | 3.4 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.3 | 2.0 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.3 | 1.4 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.3 | 0.5 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.3 | 2.6 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.3 | 1.6 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.3 | 3.7 | GO:0048148 | behavioral response to cocaine(GO:0048148) |
| 0.3 | 1.3 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.3 | 2.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.3 | 2.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 3.5 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.2 | 0.7 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.2 | 1.5 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.2 | 3.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 4.4 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.2 | 1.5 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.2 | 3.9 | GO:0071549 | response to dexamethasone(GO:0071548) cellular response to dexamethasone stimulus(GO:0071549) |
| 0.2 | 1.4 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.2 | 0.9 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.2 | 3.1 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.2 | 11.1 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.2 | 1.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.2 | 0.7 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.2 | 11.4 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.2 | 1.1 | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) |
| 0.2 | 0.9 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.2 | 3.6 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.2 | 2.2 | GO:0046959 | habituation(GO:0046959) |
| 0.2 | 1.1 | GO:0060268 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of respiratory burst(GO:0060268) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.2 | 1.3 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.2 | 0.6 | GO:0021852 | pyramidal neuron migration(GO:0021852) |
| 0.2 | 1.5 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.2 | 1.0 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.2 | 2.8 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.2 | 0.6 | GO:0035937 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.2 | 0.6 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.2 | 0.4 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.2 | 3.0 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.2 | 0.8 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.2 | 1.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 0.8 | GO:0015871 | choline transport(GO:0015871) |
| 0.2 | 1.5 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.2 | 0.8 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.2 | 2.1 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.2 | 1.3 | GO:1902302 | regulation of potassium ion export(GO:1902302) |
| 0.2 | 0.6 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.2 | 1.5 | GO:0036506 | maintenance of unfolded protein(GO:0036506) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.2 | 1.7 | GO:0034308 | primary alcohol metabolic process(GO:0034308) |
| 0.2 | 1.5 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.2 | 1.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.2 | 1.5 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.2 | 0.7 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.2 | 0.4 | GO:0001757 | somite specification(GO:0001757) |
| 0.2 | 2.3 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.2 | 1.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.2 | 2.7 | GO:0010954 | positive regulation of protein processing(GO:0010954) |
| 0.2 | 0.5 | GO:0034035 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.2 | 1.3 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.2 | 0.8 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.2 | 1.6 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.2 | 1.3 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.2 | 0.5 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.2 | 0.3 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.2 | 0.8 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.2 | 1.1 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.2 | 0.6 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 | 1.6 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 2.5 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 1.0 | GO:0071806 | intracellular protein transmembrane transport(GO:0065002) protein transmembrane transport(GO:0071806) |
| 0.1 | 5.3 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.1 | 3.4 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.1 | 5.8 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 2.7 | GO:0043496 | regulation of protein homodimerization activity(GO:0043496) |
| 0.1 | 0.7 | GO:0048793 | pronephros development(GO:0048793) |
| 0.1 | 1.5 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 1.1 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 1.2 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.1 | 1.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 0.9 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 0.5 | GO:0071499 | response to laminar fluid shear stress(GO:0034616) cellular response to laminar fluid shear stress(GO:0071499) |
| 0.1 | 0.2 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) regulation of locomotion involved in locomotory behavior(GO:0090325) negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.1 | 0.5 | GO:0009212 | dTTP biosynthetic process(GO:0006235) deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.1 | 0.8 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 | 1.4 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 1.2 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.1 | 2.2 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 0.6 | GO:0070162 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 0.6 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.1 | 1.2 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.1 | 2.7 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 0.7 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.1 | 0.7 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 | 3.9 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 0.3 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.1 | 1.9 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.1 | 1.4 | GO:0048520 | positive regulation of behavior(GO:0048520) |
| 0.1 | 1.3 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.1 | 4.0 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 2.2 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.1 | 1.3 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 2.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 5.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 2.6 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 0.7 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.1 | 3.8 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.1 | 0.9 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.1 | 4.6 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 1.3 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 0.2 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.1 | 0.8 | GO:0031054 | pre-miRNA processing(GO:0031054) |
| 0.1 | 0.7 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.1 | 0.5 | GO:0015960 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.1 | 0.5 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 2.9 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.1 | 8.6 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 2.3 | GO:0033003 | regulation of mast cell activation(GO:0033003) |
| 0.1 | 0.9 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.1 | 0.5 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 | 10.8 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.1 | 1.7 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 8.1 | GO:0048167 | regulation of synaptic plasticity(GO:0048167) |
| 0.1 | 0.8 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 1.7 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.1 | 0.2 | GO:0072236 | renal inner medulla development(GO:0072053) renal outer medulla development(GO:0072054) metanephric loop of Henle development(GO:0072236) |
| 0.1 | 4.2 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.1 | 1.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 1.0 | GO:1990173 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.1 | 1.2 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 0.2 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.1 | 1.0 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.1 | 5.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 4.7 | GO:0017157 | regulation of exocytosis(GO:0017157) |
| 0.1 | 0.3 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.1 | 0.1 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.1 | 1.5 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 3.5 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.1 | 0.4 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.1 | 2.0 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.1 | 0.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 0.8 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.1 | 0.6 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 1.3 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 2.8 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 3.1 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 0.7 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 1.7 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 0.5 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.1 | 2.3 | GO:0015807 | L-amino acid transport(GO:0015807) |
| 0.1 | 0.2 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.1 | 1.2 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.1 | 1.5 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.1 | 1.2 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 0.1 | GO:2000273 | positive regulation of receptor activity(GO:2000273) |
| 0.1 | 0.1 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.1 | 0.6 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.1 | 0.4 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.1 | 0.3 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 2.2 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.1 | 0.4 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 1.1 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 1.7 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.1 | 2.2 | GO:0006110 | regulation of glycolytic process(GO:0006110) |
| 0.1 | 1.2 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.1 | 0.1 | GO:0002842 | positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.1 | 1.2 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.1 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 0.7 | GO:1900153 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.1 | 0.5 | GO:0072201 | negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.1 | 0.7 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.1 | 0.3 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 0.5 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.1 | 0.2 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.1 | 0.1 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.1 | 0.3 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.1 | 0.7 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.1 | 1.2 | GO:0030819 | positive regulation of cAMP biosynthetic process(GO:0030819) |
| 0.1 | 0.2 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.1 | 0.3 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.1 | 0.7 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.2 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.2 | GO:2001184 | regulation of interleukin-12 secretion(GO:2001182) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 2.4 | GO:0098840 | protein transport along microtubule(GO:0098840) |
| 0.0 | 0.2 | GO:1900110 | negative regulation of histone H3-K9 dimethylation(GO:1900110) |
| 0.0 | 0.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 3.2 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.8 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.7 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.4 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.5 | GO:0002176 | male germ cell proliferation(GO:0002176) |
| 0.0 | 0.2 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 3.8 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.1 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.0 | 0.2 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.3 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.4 | GO:0002329 | immature B cell differentiation(GO:0002327) pre-B cell differentiation(GO:0002329) |
| 0.0 | 2.0 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.4 | GO:0002209 | behavioral fear response(GO:0001662) behavioral defense response(GO:0002209) fear response(GO:0042596) |
| 0.0 | 0.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.2 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.4 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.0 | 0.3 | GO:0001916 | positive regulation of T cell mediated cytotoxicity(GO:0001916) |
| 0.0 | 0.1 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
| 0.0 | 0.2 | GO:0009125 | nucleoside monophosphate catabolic process(GO:0009125) |
| 0.0 | 0.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.1 | GO:0000050 | urea cycle(GO:0000050) urea metabolic process(GO:0019627) cellular response to lithium ion(GO:0071285) |
| 0.0 | 1.5 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 1.3 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.1 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.1 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.5 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.0 | 0.2 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.0 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 1.4 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.0 | GO:1905037 | autophagosome organization(GO:1905037) |
| 0.0 | 0.3 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.7 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 0.1 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.1 | GO:0023021 | termination of signal transduction(GO:0023021) termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 | 0.2 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.2 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 56.3 | GO:0045298 | tubulin complex(GO:0045298) |
| 4.7 | 18.9 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 4.2 | 25.3 | GO:0008091 | spectrin(GO:0008091) |
| 2.9 | 8.6 | GO:0098830 | presynaptic endosome(GO:0098830) |
| 2.5 | 12.7 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 2.5 | 9.8 | GO:0044307 | dendritic branch(GO:0044307) |
| 2.3 | 6.8 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 2.0 | 6.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 1.8 | 7.3 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 1.8 | 9.1 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 1.7 | 10.4 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 1.6 | 3.3 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 1.5 | 28.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 1.4 | 15.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 1.3 | 3.8 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 1.2 | 3.6 | GO:0097637 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 1.2 | 3.5 | GO:0072534 | perineuronal net(GO:0072534) |
| 1.1 | 14.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 1.0 | 4.0 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 1.0 | 2.9 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 1.0 | 5.8 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.9 | 2.6 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.9 | 14.8 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.8 | 4.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.8 | 27.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.8 | 7.9 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.8 | 5.5 | GO:0070695 | FHF complex(GO:0070695) |
| 0.8 | 4.5 | GO:0045179 | apical cortex(GO:0045179) |
| 0.7 | 2.2 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.7 | 2.7 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.7 | 17.9 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.7 | 9.9 | GO:0043196 | varicosity(GO:0043196) |
| 0.6 | 8.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.6 | 1.8 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.6 | 12.1 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.6 | 2.8 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.6 | 2.8 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.5 | 10.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.5 | 1.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.5 | 7.0 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.5 | 1.0 | GO:0098888 | extrinsic component of presynaptic membrane(GO:0098888) |
| 0.5 | 18.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.5 | 1.5 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.5 | 4.9 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.5 | 6.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.5 | 30.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.4 | 1.3 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.4 | 1.3 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.4 | 2.6 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.4 | 5.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.4 | 2.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.4 | 1.2 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.4 | 3.7 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.4 | 3.2 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.4 | 5.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.4 | 3.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.4 | 1.5 | GO:0043293 | apoptosome(GO:0043293) |
| 0.4 | 2.5 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.4 | 2.9 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.4 | 8.4 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.4 | 2.8 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.3 | 5.6 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.3 | 1.7 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.3 | 1.0 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.3 | 4.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.3 | 12.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.3 | 5.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.3 | 2.0 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.3 | 2.5 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.3 | 0.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 0.9 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.3 | 7.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.3 | 2.9 | GO:0005916 | fascia adherens(GO:0005916) catenin complex(GO:0016342) desmosome(GO:0030057) catenin-TCF7L2 complex(GO:0071664) |
| 0.3 | 2.0 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.3 | 0.9 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.3 | 11.2 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.3 | 5.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.3 | 1.4 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.3 | 1.9 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.3 | 1.0 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.2 | 2.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.2 | 0.7 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.2 | 16.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 12.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.2 | 4.8 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.2 | 3.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 2.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 1.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.2 | 8.2 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.2 | 9.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 1.5 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.2 | 14.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.2 | 2.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.2 | 1.9 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.2 | 1.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 8.1 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.2 | 4.8 | GO:0005921 | gap junction(GO:0005921) |
| 0.2 | 6.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 0.6 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.2 | 9.3 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.2 | 0.5 | GO:0098984 | neuron to neuron synapse(GO:0098984) |
| 0.2 | 10.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.2 | 2.7 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 0.2 | GO:0097361 | CIA complex(GO:0097361) |
| 0.2 | 0.9 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.2 | 8.7 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.2 | 3.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 0.5 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.2 | 1.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.2 | 1.8 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.2 | 1.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.2 | 6.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.2 | 2.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.2 | 5.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.2 | 2.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 3.4 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.2 | 1.7 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) endocytic vesicle membrane(GO:0030666) clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.2 | 23.7 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 11.0 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 1.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 0.6 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 33.1 | GO:0099572 | postsynaptic specialization(GO:0099572) |
| 0.1 | 4.6 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 1.7 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.8 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 0.6 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 3.9 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.1 | 1.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.6 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.5 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.1 | 1.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 2.5 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 4.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 12.5 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.1 | 2.5 | GO:0032589 | neuron projection membrane(GO:0032589) |
| 0.1 | 0.6 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 5.8 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 0.5 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 2.5 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 1.0 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 1.5 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 1.1 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.1 | 0.9 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 1.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 1.0 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 5.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 16.0 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 2.7 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.5 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 0.5 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.3 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.1 | 1.7 | GO:0044297 | cell body(GO:0044297) |
| 0.1 | 14.0 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 2.5 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 1.6 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 2.2 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 1.0 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 0.9 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 0.4 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.1 | 0.4 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.3 | GO:0071817 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) MMXD complex(GO:0071817) |
| 0.1 | 1.8 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 2.4 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.1 | 2.6 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.1 | 1.3 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.0 | 0.4 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.5 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.2 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.3 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 1.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 3.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.6 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.7 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.0 | 0.9 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 2.1 | GO:0031301 | integral component of organelle membrane(GO:0031301) |
| 0.0 | 3.8 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.7 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.8 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 1.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 1.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 61.9 | GO:0031224 | intrinsic component of membrane(GO:0031224) |
| 0.0 | 0.1 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 2.4 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 4.2 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.8 | GO:0098562 | cytoplasmic side of membrane(GO:0098562) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:0000346 | transcription export complex(GO:0000346) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.0 | 56.3 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 3.8 | 18.9 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 3.6 | 21.9 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 3.4 | 10.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 3.4 | 16.9 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 2.2 | 19.8 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 2.0 | 10.0 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 2.0 | 6.0 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 1.9 | 11.7 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 1.9 | 5.8 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 1.9 | 9.3 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 1.8 | 7.3 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 1.7 | 5.0 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) epinephrine binding(GO:0051379) |
| 1.7 | 18.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 1.6 | 9.8 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 1.6 | 15.8 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 1.5 | 5.9 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 1.4 | 4.1 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 1.4 | 32.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 1.3 | 1.3 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 1.3 | 5.1 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 1.3 | 6.3 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 1.2 | 31.8 | GO:0043274 | phospholipase binding(GO:0043274) |
| 1.2 | 7.3 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 1.2 | 10.8 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 1.2 | 7.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 1.1 | 3.2 | GO:0019107 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 1.0 | 3.0 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 1.0 | 1.0 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 1.0 | 5.8 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.9 | 7.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.9 | 2.8 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.9 | 5.3 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.9 | 2.7 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.8 | 2.5 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.8 | 3.4 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.8 | 5.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.8 | 10.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.8 | 4.0 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.8 | 3.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.8 | 10.8 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.8 | 7.7 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.8 | 4.6 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.7 | 9.7 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.7 | 8.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.7 | 5.9 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.7 | 2.2 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.7 | 2.8 | GO:0002135 | CTP binding(GO:0002135) |
| 0.7 | 17.0 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.7 | 2.7 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.7 | 2.6 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.7 | 6.6 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.7 | 2.6 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.6 | 1.3 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.6 | 14.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.6 | 3.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.6 | 2.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.6 | 3.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.6 | 10.6 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.6 | 3.6 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.6 | 4.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.6 | 1.7 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.6 | 1.7 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.6 | 2.2 | GO:0036435 | deubiquitinase activator activity(GO:0035800) K48-linked polyubiquitin binding(GO:0036435) |
| 0.6 | 2.2 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.5 | 4.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.5 | 8.0 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.5 | 2.7 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.5 | 3.6 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.5 | 5.9 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.5 | 1.9 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.5 | 1.9 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.5 | 2.8 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.5 | 1.4 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) oncostatin-M receptor activity(GO:0004924) |
| 0.5 | 3.6 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.4 | 2.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.4 | 2.2 | GO:2001070 | starch binding(GO:2001070) |
| 0.4 | 3.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.4 | 1.3 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.4 | 2.2 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.4 | 1.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.4 | 3.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.4 | 2.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.4 | 7.4 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.4 | 2.5 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.4 | 2.9 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.4 | 2.4 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.4 | 9.3 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.4 | 1.2 | GO:0031779 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.4 | 1.2 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.4 | 11.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.4 | 1.5 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.4 | 2.6 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.4 | 3.7 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.4 | 1.8 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.4 | 1.5 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.4 | 7.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.4 | 1.8 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.3 | 9.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.3 | 4.9 | GO:0031005 | filamin binding(GO:0031005) |
| 0.3 | 14.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.3 | 3.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.3 | 2.7 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.3 | 1.0 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.3 | 14.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.3 | 2.0 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.3 | 2.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.3 | 1.0 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.3 | 1.0 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.3 | 4.2 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.3 | 3.8 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.3 | 6.4 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.3 | 3.2 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.3 | 2.5 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.3 | 1.2 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.3 | 6.5 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.3 | 1.8 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.3 | 2.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.3 | 1.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.3 | 0.9 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.3 | 5.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.3 | 1.5 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.3 | 9.9 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.3 | 1.5 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.3 | 5.2 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.3 | 3.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.3 | 10.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.3 | 1.7 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.3 | 11.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.3 | 3.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.3 | 4.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.3 | 12.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.3 | 2.8 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.3 | 1.9 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.3 | 6.8 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.3 | 2.7 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.3 | 1.3 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.3 | 11.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.3 | 3.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.3 | 1.3 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.2 | 1.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.2 | 2.9 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 3.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 3.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.2 | 7.6 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.2 | 1.2 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.2 | 3.7 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.2 | 3.7 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.2 | 1.8 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.2 | 12.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.2 | 0.9 | GO:0008061 | chitin binding(GO:0008061) |
| 0.2 | 2.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.2 | 1.3 | GO:0016453 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.2 | 3.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 6.9 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.2 | 2.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.2 | 1.0 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.2 | 0.6 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 1.0 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.2 | 0.8 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.2 | 0.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.2 | 1.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 19.5 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.2 | 8.3 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.2 | 2.7 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.2 | 1.8 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 0.5 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.2 | 3.2 | GO:0017022 | myosin binding(GO:0017022) |
| 0.2 | 0.7 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.2 | 3.5 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 0.7 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) reelin receptor activity(GO:0038025) |
| 0.2 | 16.0 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.2 | 0.3 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.2 | 7.0 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.2 | 0.7 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.2 | 0.8 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.2 | 0.8 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.2 | 1.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.2 | 6.8 | GO:0032934 | cholesterol binding(GO:0015485) sterol binding(GO:0032934) |
| 0.2 | 3.9 | GO:0071617 | lysophospholipid acyltransferase activity(GO:0071617) |
| 0.2 | 5.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.2 | 0.9 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.2 | 1.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 1.6 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 1.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.9 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.1 | 6.3 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 0.7 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.7 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.1 | 1.9 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 1.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 19.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 2.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 4.9 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 0.9 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.1 | 0.9 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 0.4 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 1.5 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.1 | 1.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 3.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.5 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.1 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.9 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 1.0 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 5.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 0.6 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 2.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.2 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.1 | 0.3 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.1 | 1.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 1.3 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 0.4 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.1 | 6.6 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.1 | 1.0 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 1.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 3.7 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 1.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 1.2 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 1.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 7.6 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 1.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 1.1 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 1.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 2.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.2 | GO:0070736 | protein-glycine ligase activity(GO:0070735) protein-glycine ligase activity, initiating(GO:0070736) |
| 0.1 | 0.4 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 1.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.6 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.1 | 5.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.4 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 3.2 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.5 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 0.1 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.1 | 4.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.6 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 0.7 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 0.2 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.1 | 1.3 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 11.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 2.9 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 1.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 4.9 | GO:0016798 | hydrolase activity, acting on glycosyl bonds(GO:0016798) |
| 0.1 | 0.6 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.1 | 0.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.3 | GO:0023023 | MHC protein complex binding(GO:0023023) |
| 0.1 | 4.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 2.1 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 0.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 1.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 3.5 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.4 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.2 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.0 | 1.1 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 0.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.7 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 2.7 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 2.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.3 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 1.0 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.7 | GO:0008234 | cysteine-type peptidase activity(GO:0008234) |
| 0.0 | 1.1 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 2.1 | GO:0015399 | primary active transmembrane transporter activity(GO:0015399) P-P-bond-hydrolysis-driven transmembrane transporter activity(GO:0015405) |
| 0.0 | 0.5 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.7 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 1.5 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.1 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.5 | GO:0015605 | organophosphate ester transmembrane transporter activity(GO:0015605) |
| 0.0 | 0.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.4 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.5 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 3.9 | GO:0005509 | calcium ion binding(GO:0005509) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 56.3 | PID_P38_GAMMA_DELTA_PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 2.2 | 37.6 | PID_LPA4_PATHWAY | LPA4-mediated signaling events |
| 1.2 | 28.1 | ST_WNT_CA2_CYCLIC_GMP_PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.4 | 7.1 | PID_IGF1_PATHWAY | IGF1 pathway |
| 0.4 | 6.4 | PID_PRL_SIGNALING_EVENTS_PATHWAY | Signaling events mediated by PRL |
| 0.3 | 5.1 | PID_PI3KCI_PATHWAY | Class I PI3K signaling events |
| 0.3 | 6.1 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.3 | 18.5 | PID_ERBB1_INTERNALIZATION_PATHWAY | Internalization of ErbB1 |
| 0.3 | 14.1 | PID_INTEGRIN_A4B1_PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.3 | 19.1 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.3 | 17.0 | ST_PHOSPHOINOSITIDE_3_KINASE_PATHWAY | PI3K Pathway |
| 0.3 | 6.3 | PID_S1P_META_PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.3 | 0.8 | PID_WNT_CANONICAL_PATHWAY | Canonical Wnt signaling pathway |
| 0.3 | 8.9 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 6.1 | PID_PI3KCI_AKT_PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.2 | 2.8 | PID_IL12_STAT4_PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 8.9 | PID_INSULIN_PATHWAY | Insulin Pathway |
| 0.2 | 6.7 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.2 | 2.9 | PID_ECADHERIN_KERATINOCYTE_PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 4.3 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.2 | 7.2 | PID_REELIN_PATHWAY | Reelin signaling pathway |
| 0.2 | 4.4 | PID_GLYPICAN_1PATHWAY | Glypican 1 network |
| 0.2 | 6.0 | PID_TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.2 | 2.9 | NABA_BASEMENT_MEMBRANES | Genes encoding structural components of basement membranes |
| 0.2 | 5.0 | SIG_BCR_SIGNALING_PATHWAY | Members of the BCR signaling pathway |
| 0.2 | 11.3 | PID_P73PATHWAY | p73 transcription factor network |
| 0.2 | 5.2 | PID_P38_ALPHA_BETA_PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.2 | 1.7 | PID_RHOA_PATHWAY | RhoA signaling pathway |
| 0.2 | 4.5 | PID_EPHA_FWDPATHWAY | EPHA forward signaling |
| 0.2 | 19.3 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 7.0 | PID_P75_NTR_PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 2.3 | PID_MAPK_TRK_PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 4.0 | PID_TRKR_PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 1.3 | PID_IL6_7_PATHWAY | IL6-mediated signaling events |
| 0.1 | 1.9 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.1 | 1.2 | PID_NFKAPPAB_CANONICAL_PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 11.9 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.2 | PID_INTEGRIN_A9B1_PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 1.4 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.1 | 1.1 | PID_P38_ALPHA_BETA_DOWNSTREAM_PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 1.4 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.1 | 0.9 | PID_NFAT_TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 2.0 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 0.9 | PID_ALPHA_SYNUCLEIN_PATHWAY | Alpha-synuclein signaling |
| 0.1 | 0.6 | PID_TRAIL_PATHWAY | TRAIL signaling pathway |
| 0.1 | 2.1 | PID_TGFBR_PATHWAY | TGF-beta receptor signaling |
| 0.1 | 0.6 | PID_ARF6_DOWNSTREAM_PATHWAY | Arf6 downstream pathway |
| 0.1 | 2.2 | ST_INTEGRIN_SIGNALING_PATHWAY | Integrin Signaling Pathway |
| 0.1 | 2.2 | PID_MTOR_4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.3 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.0 | 1.7 | PID_P53_REGULATION_PATHWAY | p53 pathway |
| 0.0 | 0.3 | ST_P38_MAPK_PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.4 | SIG_CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.4 | PID_DNA_PK_PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.8 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.1 | PID_INTEGRIN3_PATHWAY | Beta3 integrin cell surface interactions |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 56.3 | REACTOME_CASPASE_MEDIATED_CLEAVAGE_OF_CYTOSKELETAL_PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 2.3 | 27.7 | REACTOME_ADENYLATE_CYCLASE_ACTIVATING_PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 1.8 | 7.0 | REACTOME_DSCAM_INTERACTIONS | Genes involved in DSCAM interactions |
| 1.8 | 15.8 | REACTOME_ACETYLCHOLINE_BINDING_AND_DOWNSTREAM_EVENTS | Genes involved in Acetylcholine Binding And Downstream Events |
| 1.6 | 40.0 | REACTOME_CREB_PHOSPHORYLATION_THROUGH_THE_ACTIVATION_OF_CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 1.5 | 7.7 | REACTOME_RAP1_SIGNALLING | Genes involved in Rap1 signalling |
| 1.2 | 14.8 | REACTOME_PKA_MEDIATED_PHOSPHORYLATION_OF_CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 1.2 | 18.9 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 1.0 | 34.9 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.8 | 2.5 | REACTOME_ADENYLATE_CYCLASE_INHIBITORY_PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.7 | 1.3 | REACTOME_ELEVATION_OF_CYTOSOLIC_CA2_LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.6 | 22.7 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.6 | 10.6 | REACTOME_HORMONE_SENSITIVE_LIPASE_HSL_MEDIATED_TRIACYLGLYCEROL_HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.6 | 13.0 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.5 | 7.1 | REACTOME_NUCLEOTIDE_LIKE_PURINERGIC_RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.5 | 11.7 | REACTOME_KERATAN_SULFATE_BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.5 | 15.3 | REACTOME_A_TETRASACCHARIDE_LINKER_SEQUENCE_IS_REQUIRED_FOR_GAG_SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.5 | 3.2 | REACTOME_THE_NLRP3_INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.4 | 10.4 | REACTOME_INSULIN_SYNTHESIS_AND_PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.4 | 6.8 | REACTOME_SIGNALING_BY_NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.4 | 8.3 | REACTOME_CRMPS_IN_SEMA3A_SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.4 | 17.0 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.4 | 5.7 | REACTOME_NEF_MEDIATED_DOWNREGULATION_OF_MHC_CLASS_I_COMPLEX_CELL_SURFACE_EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.4 | 3.2 | REACTOME_MEMBRANE_BINDING_AND_TARGETTING_OF_GAG_PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.4 | 13.2 | REACTOME_FATTY_ACYL_COA_BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.4 | 12.3 | REACTOME_ERK_MAPK_TARGETS | Genes involved in ERK/MAPK targets |
| 0.4 | 3.4 | REACTOME_NRIF_SIGNALS_CELL_DEATH_FROM_THE_NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.3 | 2.8 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PI | Genes involved in Acyl chain remodelling of PI |
| 0.3 | 8.6 | REACTOME_DOWNREGULATION_OF_TGF_BETA_RECEPTOR_SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.3 | 3.1 | REACTOME_ABACAVIR_TRANSPORT_AND_METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.3 | 3.5 | REACTOME_ROLE_OF_DCC_IN_REGULATING_APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.3 | 5.8 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.3 | 2.9 | REACTOME_CHYLOMICRON_MEDIATED_LIPID_TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 9.0 | REACTOME_ANTIGEN_ACTIVATES_B_CELL_RECEPTOR_LEADING_TO_GENERATION_OF_SECOND_MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.3 | 6.5 | REACTOME_TRAFFICKING_OF_GLUR2_CONTAINING_AMPA_RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.3 | 4.9 | REACTOME_ANTIGEN_PRESENTATION_FOLDING_ASSEMBLY_AND_PEPTIDE_LOADING_OF_CLASS_I_MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.3 | 4.8 | REACTOME_GABA_SYNTHESIS_RELEASE_REUPTAKE_AND_DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.3 | 2.4 | REACTOME_PLATELET_SENSITIZATION_BY_LDL | Genes involved in Platelet sensitization by LDL |
| 0.3 | 2.7 | REACTOME_SEMA3A_PAK_DEPENDENT_AXON_REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.3 | 2.5 | REACTOME_NA_CL_DEPENDENT_NEUROTRANSMITTER_TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 1.0 | REACTOME_THROMBOXANE_SIGNALLING_THROUGH_TP_RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.2 | 9.3 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 2.4 | REACTOME_ACTIVATION_OF_CHAPERONES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.2 | 2.8 | REACTOME_SLBP_DEPENDENT_PROCESSING_OF_REPLICATION_DEPENDENT_HISTONE_PRE_MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 3.9 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION_IN_THE_MEDIAL_TRANS_GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.2 | 2.1 | REACTOME_PROTEOLYTIC_CLEAVAGE_OF_SNARE_COMPLEX_PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 2.7 | REACTOME_CYTOCHROME_P450_ARRANGED_BY_SUBSTRATE_TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.2 | 0.4 | REACTOME_SHC_MEDIATED_SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.2 | 2.5 | REACTOME_SOS_MEDIATED_SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.2 | 5.8 | REACTOME_G_PROTEIN_ACTIVATION | Genes involved in G-protein activation |
| 0.2 | 2.2 | REACTOME_SYNTHESIS_OF_PE | Genes involved in Synthesis of PE |
| 0.2 | 2.9 | REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_GLUCAGON_LIKE_PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.2 | 2.2 | REACTOME_REGULATION_OF_RHEB_GTPASE_ACTIVITY_BY_AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.2 | 6.3 | REACTOME_POTASSIUM_CHANNELS | Genes involved in Potassium Channels |
| 0.2 | 0.9 | REACTOME_GRB2_SOS_PROVIDES_LINKAGE_TO_MAPK_SIGNALING_FOR_INTERGRINS_ | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.2 | 7.2 | REACTOME_NETRIN1_SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 3.0 | REACTOME_ABCA_TRANSPORTERS_IN_LIPID_HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 2.9 | REACTOME_BASIGIN_INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 8.4 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.2 | REACTOME_RECRUITMENT_OF_NUMA_TO_MITOTIC_CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 2.2 | REACTOME_POST_CHAPERONIN_TUBULIN_FOLDING_PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.6 | REACTOME_CHONDROITIN_SULFATE_BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 5.0 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 2.4 | REACTOME_METABOLISM_OF_PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 3.7 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 0.4 | REACTOME_ANDROGEN_BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 2.3 | REACTOME_AMINO_ACID_TRANSPORT_ACROSS_THE_PLASMA_MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 0.8 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PC | Genes involved in Acyl chain remodelling of PC |
| 0.1 | 8.9 | REACTOME_P75_NTR_RECEPTOR_MEDIATED_SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 0.9 | REACTOME_ALPHA_LINOLENIC_ACID_ALA_METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 1.2 | REACTOME_ACTIVATION_OF_BH3_ONLY_PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 1.0 | REACTOME_TGF_BETA_RECEPTOR_SIGNALING_IN_EMT_EPITHELIAL_TO_MESENCHYMAL_TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.9 | REACTOME_CYTOSOLIC_TRNA_AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 1.2 | REACTOME_JNK_C_JUN_KINASES_PHOSPHORYLATION_AND_ACTIVATION_MEDIATED_BY_ACTIVATED_HUMAN_TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 1.6 | REACTOME_METAL_ION_SLC_TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 5.8 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 1.5 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 0.8 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 1.1 | REACTOME_EGFR_DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 0.6 | REACTOME_VEGF_LIGAND_RECEPTOR_INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.1 | REACTOME_PERK_REGULATED_GENE_EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.1 | 0.8 | REACTOME_INTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.8 | REACTOME_SIGNALING_BY_NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 3.9 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.4 | REACTOME_GLYCOSPHINGOLIPID_METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.9 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 2.2 | REACTOME_METABOLISM_OF_VITAMINS_AND_COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.7 | REACTOME_ANTIVIRAL_MECHANISM_BY_IFN_STIMULATED_GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.7 | REACTOME_INSULIN_RECEPTOR_RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 1.0 | REACTOME_TRANSPORT_OF_GLUCOSE_AND_OTHER_SUGARS_BILE_SALTS_AND_ORGANIC_ACIDS_METAL_IONS_AND_AMINE_COMPOUNDS | Genes involved in Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds |
| 0.0 | 0.3 | REACTOME_G_BETA_GAMMA_SIGNALLING_THROUGH_PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 1.0 | REACTOME_O_LINKED_GLYCOSYLATION_OF_MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.2 | REACTOME_TRANSPORT_OF_VITAMINS_NUCLEOSIDES_AND_RELATED_MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.2 | REACTOME_INTEGRATION_OF_ENERGY_METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.3 | REACTOME_PYRIMIDINE_CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 2.0 | REACTOME_NEURONAL_SYSTEM | Genes involved in Neuronal System |
| 0.0 | 0.5 | REACTOME_TRANSPORT_OF_RIBONUCLEOPROTEINS_INTO_THE_HOST_NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.4 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.1 | REACTOME_CYTOSOLIC_SULFONATION_OF_SMALL_MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.8 | REACTOME_SIGNALING_BY_EGFR_IN_CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.4 | REACTOME_SYNTHESIS_OF_GLYCOSYLPHOSPHATIDYLINOSITOL_GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.7 | REACTOME_ER_PHAGOSOME_PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.1 | REACTOME_SIGNAL_REGULATORY_PROTEIN_SIRP_FAMILY_INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |