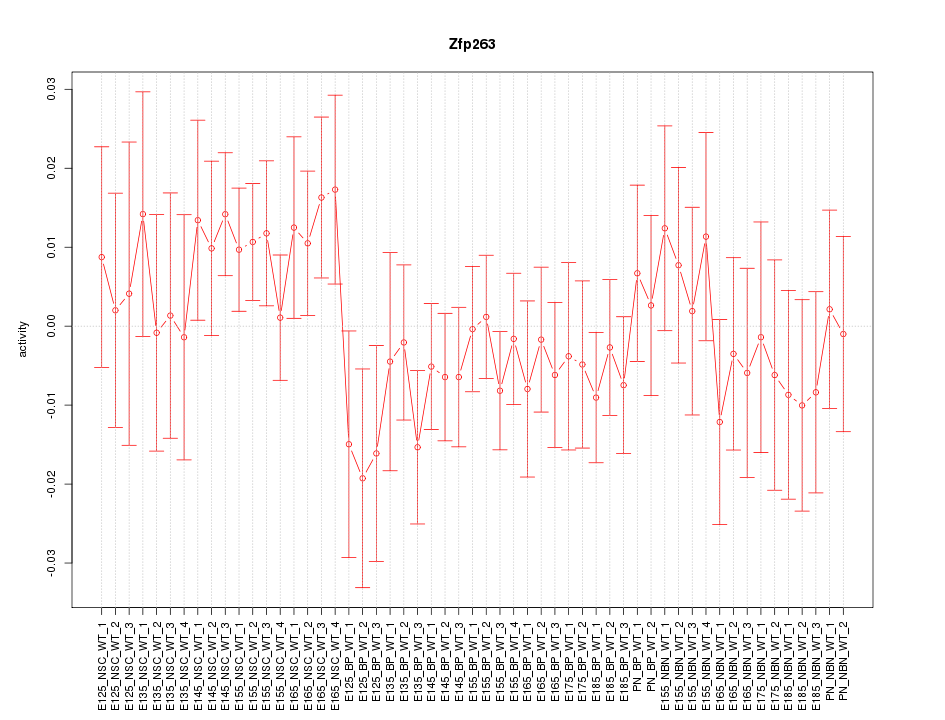
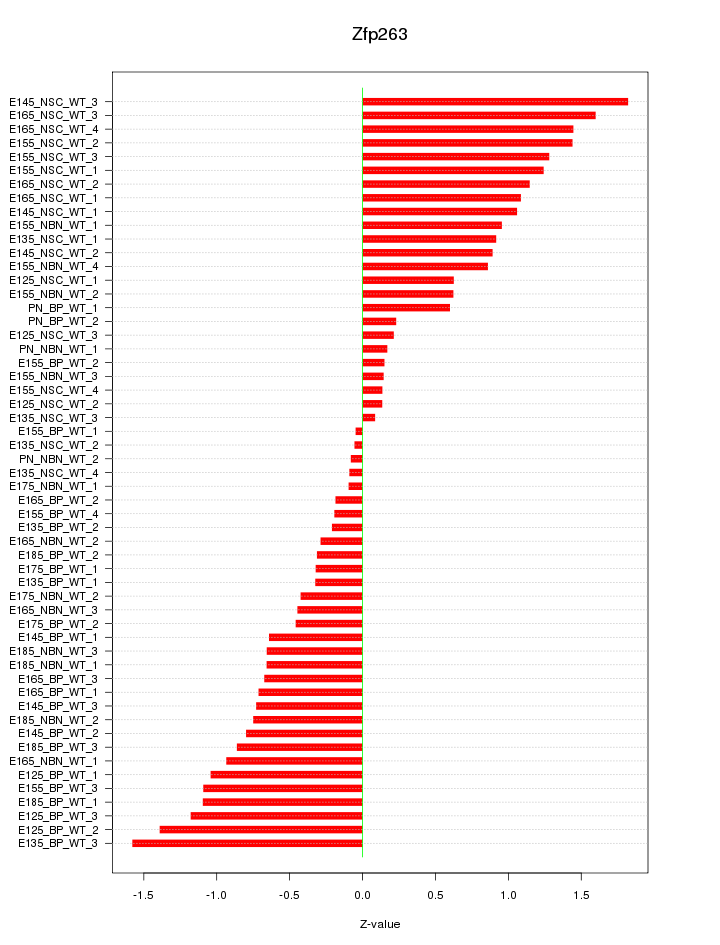
Motif ID: Zfp263
Z-value: 0.826
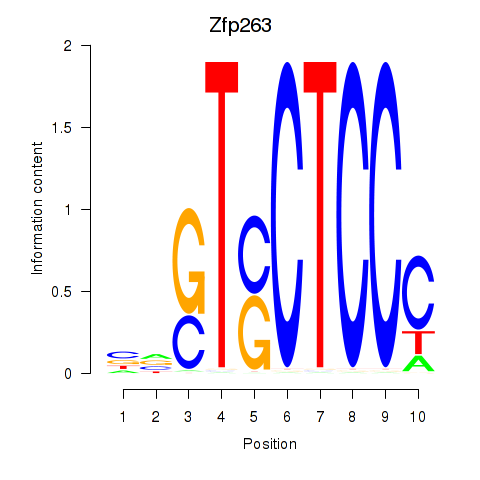
Transcription factors associated with Zfp263:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Zfp263 | ENSMUSG00000022529.5 | Zfp263 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp263 | mm10_v2_chr16_+_3744089_3744145 | 0.00 | 9.8e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.6 | GO:0060166 | olfactory pit development(GO:0060166) |
| 2.3 | 7.0 | GO:0072223 | metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 2.1 | 4.1 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 1.6 | 4.7 | GO:1904956 | dermatome development(GO:0061054) sclerotome development(GO:0061056) regulation of dermatome development(GO:0061183) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 1.5 | 4.6 | GO:0061075 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 1.2 | 3.6 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.8 | 3.8 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.7 | 2.1 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.5 | 2.7 | GO:0015817 | glutamine transport(GO:0006868) histidine transport(GO:0015817) cellular response to potassium ion starvation(GO:0051365) |
| 0.5 | 5.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.5 | 3.0 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.5 | 1.4 | GO:0038044 | negative regulation of receptor biosynthetic process(GO:0010871) transforming growth factor-beta secretion(GO:0038044) |
| 0.5 | 7.9 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.4 | 1.7 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.4 | 1.2 | GO:0043379 | memory T cell differentiation(GO:0043379) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.4 | 1.2 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.4 | 1.2 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.4 | 1.1 | GO:0097360 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.3 | 1.3 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.3 | 1.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.3 | 1.2 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) |
| 0.3 | 3.9 | GO:0046643 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.3 | 0.6 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.3 | 0.9 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.3 | 4.3 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.3 | 0.3 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.3 | 2.2 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.3 | 0.8 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.3 | 6.0 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.3 | 0.8 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.2 | 4.8 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.2 | 2.6 | GO:0042473 | outer ear morphogenesis(GO:0042473) |
| 0.2 | 2.3 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.2 | 0.9 | GO:0070384 | Harderian gland development(GO:0070384) |
| 0.2 | 0.4 | GO:0060126 | somatotropin secreting cell differentiation(GO:0060126) |
| 0.2 | 1.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.2 | 0.7 | GO:0090194 | negative regulation of glomerular mesangial cell proliferation(GO:0072125) negative regulation of glomerulus development(GO:0090194) |
| 0.2 | 2.8 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.2 | 0.6 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.2 | 1.4 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.2 | 0.4 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.2 | 0.3 | GO:0097168 | mesenchymal stem cell proliferation(GO:0097168) |
| 0.2 | 0.2 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.2 | 0.5 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.2 | 1.1 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.2 | 1.2 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.2 | 0.6 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.2 | 0.6 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.1 | 0.4 | GO:0035864 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.1 | 0.4 | GO:0035624 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.1 | 1.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.6 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 | 0.9 | GO:0032754 | positive regulation of interleukin-5 production(GO:0032754) |
| 0.1 | 0.3 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.1 | 0.5 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.1 | 1.3 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 0.4 | GO:0002030 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) |
| 0.1 | 0.6 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 | 0.9 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.8 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 | 2.7 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 2.9 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 0.6 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 | 0.4 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.1 | 0.4 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.1 | 0.3 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) regulation of connective tissue replacement(GO:1905203) |
| 0.1 | 0.4 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.1 | 0.4 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.1 | 0.3 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.1 | 2.4 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.1 | 0.9 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 0.5 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 0.6 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 1.5 | GO:0001975 | response to amphetamine(GO:0001975) response to amine(GO:0014075) |
| 0.1 | 1.0 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.1 | 0.5 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 0.2 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.1 | 0.3 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 | 0.1 | GO:0060994 | regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) |
| 0.1 | 0.3 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.1 | 0.3 | GO:0090237 | regulation of arachidonic acid secretion(GO:0090237) positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.1 | 2.0 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.1 | 0.7 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.5 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.4 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.4 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.2 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 | 0.4 | GO:0061009 | common bile duct development(GO:0061009) |
| 0.1 | 1.3 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.1 | 0.1 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 0.2 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 0.1 | 0.1 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.1 | 0.2 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 0.5 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.1 | 0.4 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.1 | 0.6 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.7 | GO:0030238 | male sex determination(GO:0030238) |
| 0.1 | 0.4 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.1 | 3.4 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.1 | 0.4 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.1 | 1.3 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 | 0.4 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 0.3 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.1 | 0.7 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.1 | 0.4 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 0.4 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.1 | 0.5 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 0.2 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.1 | 1.1 | GO:0060746 | parental behavior(GO:0060746) |
| 0.1 | 0.8 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 0.8 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 0.4 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.3 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.1 | 0.4 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) |
| 0.1 | 0.3 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 0.1 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.1 | 0.2 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.0 | 0.6 | GO:0060068 | vagina development(GO:0060068) |
| 0.0 | 0.4 | GO:0070813 | hydrogen sulfide metabolic process(GO:0070813) |
| 0.0 | 0.1 | GO:0060686 | regulation of prostatic bud formation(GO:0060685) negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 | 0.3 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.1 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.3 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.8 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.0 | 0.7 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 1.2 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.4 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 1.0 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 0.5 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.9 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.3 | GO:0060850 | regulation of transcription involved in cell fate commitment(GO:0060850) |
| 0.0 | 1.8 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 1.3 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.2 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.6 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.2 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 1.7 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 | 0.7 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.0 | 0.5 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 0.2 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.6 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.1 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.0 | 0.2 | GO:0071630 | trophectodermal cellular morphogenesis(GO:0001831) nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.1 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.3 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.6 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 1.0 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.1 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.2 | GO:0051608 | histamine transport(GO:0051608) |
| 0.0 | 0.4 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.1 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.0 | 0.2 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.1 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.2 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.1 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0003356 | regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) |
| 0.0 | 0.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.7 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.1 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 | 2.0 | GO:0050679 | positive regulation of epithelial cell proliferation(GO:0050679) |
| 0.0 | 0.5 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.1 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 1.1 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.2 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.3 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.2 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.0 | 0.1 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 1.0 | GO:0045638 | negative regulation of myeloid cell differentiation(GO:0045638) |
| 0.0 | 0.1 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.6 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.8 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.1 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.0 | 0.4 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.2 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.6 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.4 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.5 | 3.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.5 | 1.4 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 0.4 | 2.0 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.3 | 2.4 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.3 | 1.2 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.3 | 0.9 | GO:0044299 | C-fiber(GO:0044299) |
| 0.2 | 0.8 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.2 | 0.9 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 0.7 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.2 | 1.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.2 | 1.9 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 1.9 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 1.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 2.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 2.3 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 0.8 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 2.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.7 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.4 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 0.4 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 0.6 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 0.7 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 0.3 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.4 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 0.7 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 0.6 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 0.4 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.4 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.6 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 1.1 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.4 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.4 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.6 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.4 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.7 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 4.6 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 0.4 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.3 | GO:0070013 | intracellular organelle lumen(GO:0070013) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 4.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 6.4 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 2.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 4.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.2 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 1.1 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 1.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.3 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.5 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 5.8 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.2 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 2.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.5 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 2.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.3 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.4 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.6 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.8 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.9 | GO:0005923 | bicellular tight junction(GO:0005923) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.1 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.9 | 2.7 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.8 | 3.3 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.6 | 5.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.6 | 3.3 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.5 | 4.7 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.5 | 3.3 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.5 | 3.7 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.5 | 5.9 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.4 | 1.9 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.4 | 1.5 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.4 | 4.0 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.3 | 1.7 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.3 | 2.3 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.3 | 1.0 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.3 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.3 | 1.4 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) neuregulin binding(GO:0038132) |
| 0.3 | 1.6 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 3.8 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 0.7 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.2 | 1.3 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.2 | 1.5 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 0.6 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.2 | 1.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.2 | 6.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.2 | 1.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 0.9 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.2 | 0.7 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.2 | 0.6 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.1 | 0.4 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 1.6 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 2.3 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 10.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.0 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 1.1 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.1 | 0.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.4 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) epinephrine binding(GO:0051379) |
| 0.1 | 1.7 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 1.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.4 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 1.8 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 3.0 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.5 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.6 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 1.2 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.1 | 0.4 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.1 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 0.7 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 0.5 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.1 | 1.0 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 3.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 1.5 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 0.5 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.1 | 1.0 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 2.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.6 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 0.8 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 0.3 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.1 | 0.6 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.6 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.7 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.4 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.6 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.8 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 2.1 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 0.8 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.2 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.1 | 0.7 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.2 | GO:0016901 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.1 | 0.2 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.1 | 0.1 | GO:0031762 | alpha-1A adrenergic receptor binding(GO:0031691) alpha-1B adrenergic receptor binding(GO:0031692) follicle-stimulating hormone receptor binding(GO:0031762) |
| 0.1 | 0.9 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 0.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.5 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 0.2 | GO:0004454 | ketohexokinase activity(GO:0004454) |
| 0.0 | 0.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.3 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 3.2 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.5 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.2 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.7 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.6 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.4 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.8 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 1.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 1.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.8 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.8 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 2.4 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) |
| 0.0 | 0.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.3 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.3 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 2.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.2 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.3 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.9 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 1.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) lipase activator activity(GO:0060229) |
| 0.0 | 0.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0050694 | galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 1.2 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.1 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.7 | PID_INTEGRIN_CS_PATHWAY | Integrin family cell surface interactions |
| 0.2 | 9.3 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 5.2 | PID_PRL_SIGNALING_EVENTS_PATHWAY | Signaling events mediated by PRL |
| 0.2 | 5.1 | PID_NECTIN_PATHWAY | Nectin adhesion pathway |
| 0.2 | 5.0 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.9 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.1 | 5.4 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.1 | 3.4 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 2.7 | SIG_PIP3_SIGNALING_IN_B_LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.5 | PID_ALK2_PATHWAY | ALK2 signaling events |
| 0.0 | 1.2 | PID_IL12_STAT4_PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.2 | PID_WNT_CANONICAL_PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 2.4 | PID_THROMBIN_PAR1_PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 1.3 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.3 | PID_RHODOPSIN_PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.7 | PID_FOXO_PATHWAY | FoxO family signaling |
| 0.0 | 1.5 | PID_RETINOIC_ACID_PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.4 | PID_REELIN_PATHWAY | Reelin signaling pathway |
| 0.0 | 1.9 | PID_HES_HEY_PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.3 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.0 | 4.2 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.2 | PID_IL8_CXCR2_PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.5 | ST_G_ALPHA_I_PATHWAY | G alpha i Pathway |
| 0.0 | 0.5 | PID_IL1_PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.6 | PID_IL2_STAT5_PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 2.3 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.9 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.2 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.3 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | PID_RB_1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.2 | PID_CIRCADIAN_PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.6 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | REACTOME_TRANSLOCATION_OF_ZAP_70_TO_IMMUNOLOGICAL_SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 5.7 | REACTOME_INTERFERON_ALPHA_BETA_SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 7.4 | REACTOME_CROSS_PRESENTATION_OF_SOLUBLE_EXOGENOUS_ANTIGENS_ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 1.5 | REACTOME_CHONDROITIN_SULFATE_BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 4.1 | REACTOME_A_TETRASACCHARIDE_LINKER_SEQUENCE_IS_REQUIRED_FOR_GAG_SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 1.4 | REACTOME_PECAM1_INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 0.7 | REACTOME_ROLE_OF_SECOND_MESSENGERS_IN_NETRIN1_SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 1.2 | REACTOME_ROLE_OF_DCC_IN_REGULATING_APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 3.1 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.5 | REACTOME_CYCLIN_A_B1_ASSOCIATED_EVENTS_DURING_G2_M_TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 2.7 | REACTOME_AMINO_ACID_TRANSPORT_ACROSS_THE_PLASMA_MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.9 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 6.0 | REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 1.7 | REACTOME_SYNTHESIS_OF_PC | Genes involved in Synthesis of PC |
| 0.1 | 1.6 | REACTOME_EFFECTS_OF_PIP2_HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 0.5 | REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 2.4 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 0.6 | REACTOME_CHYLOMICRON_MEDIATED_LIPID_TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.2 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.7 | REACTOME_DEGRADATION_OF_THE_EXTRACELLULAR_MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.7 | REACTOME_SYNTHESIS_SECRETION_AND_DEACYLATION_OF_GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.8 | REACTOME_INHIBITION_OF_THE_PROTEOLYTIC_ACTIVITY_OF_APC_C_REQUIRED_FOR_THE_ONSET_OF_ANAPHASE_BY_MITOTIC_SPINDLE_CHECKPOINT_COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.7 | REACTOME_PROCESSING_OF_INTRONLESS_PRE_MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 1.2 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.0 | 2.3 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.3 | REACTOME_SMAD2_SMAD3_SMAD4_HETEROTRIMER_REGULATES_TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 1.1 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME_NA_CL_DEPENDENT_NEUROTRANSMITTER_TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.1 | REACTOME_DAG_AND_IP3_SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.2 | REACTOME_ORGANIC_CATION_ANION_ZWITTERION_TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.4 | REACTOME_CDC6_ASSOCIATION_WITH_THE_ORC_ORIGIN_COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.2 | REACTOME_JNK_C_JUN_KINASES_PHOSPHORYLATION_AND_ACTIVATION_MEDIATED_BY_ACTIVATED_HUMAN_TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 1.9 | REACTOME_RESPONSE_TO_ELEVATED_PLATELET_CYTOSOLIC_CA2_ | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.3 | REACTOME_AMINE_LIGAND_BINDING_RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.4 | REACTOME_GPVI_MEDIATED_ACTIVATION_CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.8 | REACTOME_IL1_SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.0 | REACTOME_EXTRACELLULAR_MATRIX_ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 1.4 | REACTOME_MEIOTIC_SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.3 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 1.2 | REACTOME_DOWNSTREAM_SIGNAL_TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.2 | REACTOME_ERKS_ARE_INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME_MTORC1_MEDIATED_SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 1.7 | REACTOME_L1CAM_INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.2 | REACTOME_METABOLISM_OF_POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.6 | REACTOME_TRAFFICKING_OF_AMPA_RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.3 | REACTOME_RNA_POL_III_CHAIN_ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.3 | REACTOME_METABOLISM_OF_STEROID_HORMONES_AND_VITAMINS_A_AND_D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.3 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME_CASPASE_MEDIATED_CLEAVAGE_OF_CYTOSKELETAL_PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.6 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |