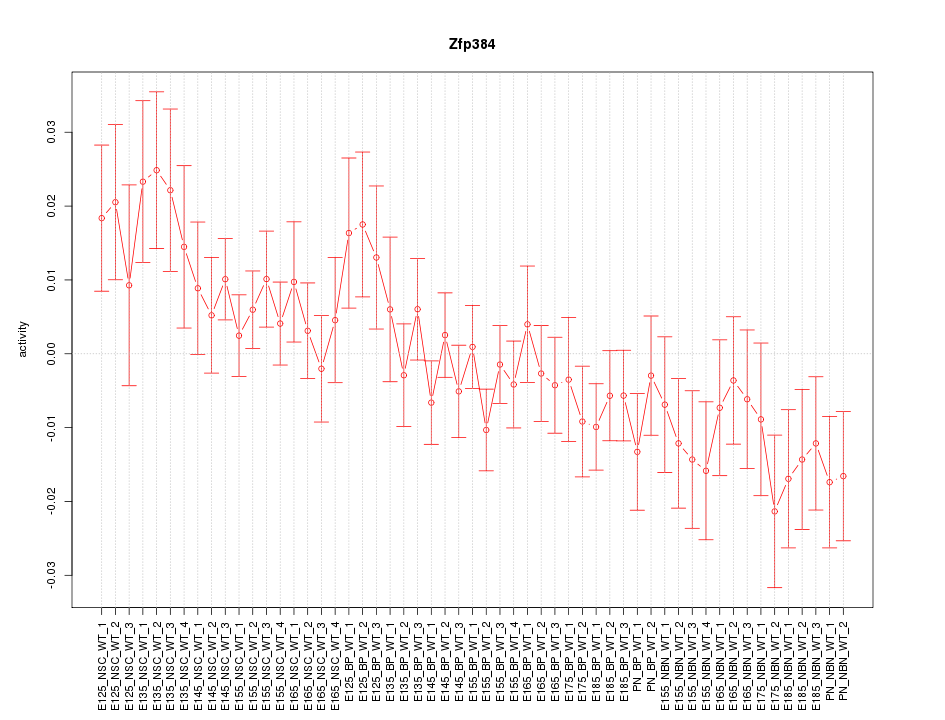
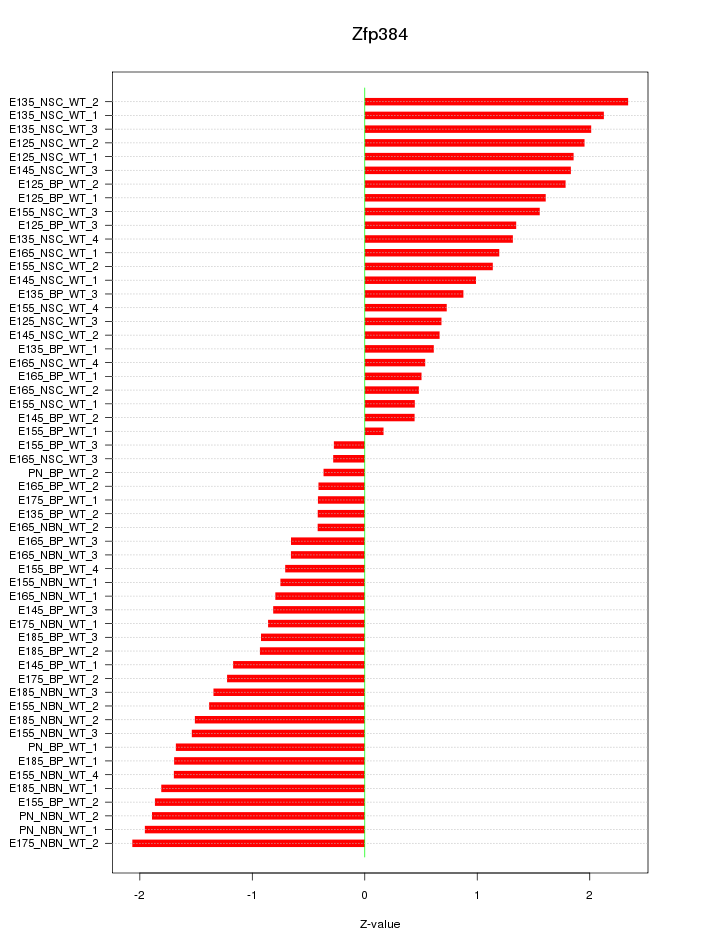
Motif ID: Zfp384
Z-value: 1.271
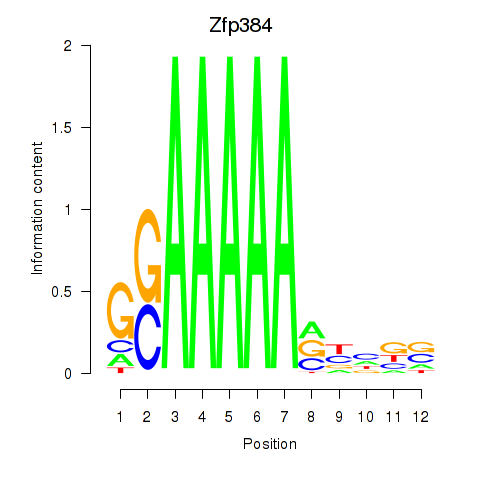
Transcription factors associated with Zfp384:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Zfp384 | ENSMUSG00000038346.12 | Zfp384 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp384 | mm10_v2_chr6_+_125009261_125009317 | -0.15 | 2.8e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.5 | GO:0097402 | neuroblast migration(GO:0097402) |
| 3.0 | 15.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 2.4 | 7.1 | GO:0003219 | atrioventricular node development(GO:0003162) cardiac right ventricle formation(GO:0003219) |
| 2.3 | 11.7 | GO:1901731 | positive regulation of platelet aggregation(GO:1901731) |
| 2.0 | 6.1 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 2.0 | 7.9 | GO:0019323 | D-ribose metabolic process(GO:0006014) pentose catabolic process(GO:0019323) |
| 1.9 | 5.8 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 1.9 | 5.6 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 1.8 | 5.3 | GO:1990523 | bone regeneration(GO:1990523) |
| 1.7 | 5.1 | GO:0060023 | soft palate development(GO:0060023) |
| 1.7 | 5.0 | GO:0021759 | globus pallidus development(GO:0021759) |
| 1.6 | 4.9 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 1.5 | 4.5 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 1.4 | 4.1 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 1.4 | 9.6 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 1.3 | 3.9 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 1.3 | 3.9 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 1.3 | 1.3 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 1.2 | 3.7 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 1.2 | 6.2 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 1.2 | 2.4 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 1.1 | 3.4 | GO:0019405 | alditol catabolic process(GO:0019405) |
| 1.1 | 3.3 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 1.1 | 5.5 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 1.1 | 2.1 | GO:0002678 | positive regulation of chronic inflammatory response(GO:0002678) |
| 1.0 | 4.2 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 1.0 | 3.0 | GO:0045819 | plasmacytoid dendritic cell activation(GO:0002270) positive regulation of glycogen catabolic process(GO:0045819) |
| 1.0 | 3.0 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 1.0 | 4.9 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.9 | 6.2 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.9 | 2.6 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.9 | 1.7 | GO:0046370 | fructose biosynthetic process(GO:0046370) |
| 0.8 | 3.4 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.8 | 1.6 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.8 | 4.0 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.8 | 2.4 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.8 | 2.4 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.8 | 3.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.8 | 2.3 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.8 | 3.0 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.8 | 8.3 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.8 | 15.8 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.8 | 7.5 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.7 | 2.9 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.7 | 2.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.7 | 0.7 | GO:0014735 | regulation of muscle atrophy(GO:0014735) |
| 0.7 | 3.5 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.7 | 7.0 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.7 | 1.4 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.7 | 2.0 | GO:0046671 | positive regulation of neuron maturation(GO:0014042) regulation of cellular pH reduction(GO:0032847) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.7 | 2.0 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.7 | 2.6 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.7 | 0.7 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.7 | 3.9 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.6 | 1.9 | GO:0003104 | positive regulation of glomerular filtration(GO:0003104) cell communication by chemical coupling(GO:0010643) |
| 0.6 | 0.6 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.6 | 4.4 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.6 | 4.9 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.6 | 6.0 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.6 | 3.6 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.6 | 1.1 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.6 | 2.8 | GO:1901526 | negative regulation of mitochondrial membrane permeability(GO:0035795) positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.6 | 4.5 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.6 | 2.2 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.5 | 1.6 | GO:0009814 | defense response, incompatible interaction(GO:0009814) |
| 0.5 | 2.2 | GO:2000484 | positive regulation of interleukin-8 secretion(GO:2000484) |
| 0.5 | 3.8 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.5 | 7.3 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.5 | 2.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.5 | 2.1 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.5 | 14.0 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.5 | 1.5 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) transforming growth factor-beta secretion(GO:0038044) |
| 0.5 | 1.5 | GO:0034334 | adherens junction maintenance(GO:0034334) intermediate filament bundle assembly(GO:0045110) maintenance of organ identity(GO:0048496) |
| 0.5 | 2.0 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.5 | 4.5 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.5 | 4.5 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.5 | 1.0 | GO:0019395 | fatty acid oxidation(GO:0019395) |
| 0.5 | 5.8 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.5 | 3.8 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.5 | 4.3 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.5 | 1.4 | GO:1904760 | myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.5 | 1.9 | GO:0051135 | positive regulation of NK T cell activation(GO:0051135) |
| 0.5 | 2.3 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.5 | 1.4 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.5 | 4.5 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.4 | 2.2 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.4 | 4.9 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.4 | 1.7 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.4 | 3.4 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.4 | 2.5 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.4 | 2.5 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.4 | 1.3 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.4 | 8.1 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.4 | 1.2 | GO:0016095 | polyprenol catabolic process(GO:0016095) terpenoid catabolic process(GO:0016115) primary alcohol catabolic process(GO:0034310) |
| 0.4 | 1.6 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.4 | 2.8 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.4 | 1.2 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.4 | 1.2 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.4 | 1.5 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.4 | 4.1 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.4 | 0.7 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.4 | 1.9 | GO:0089700 | protein kinase D signaling(GO:0089700) positive regulation of histone deacetylase activity(GO:1901727) |
| 0.4 | 1.1 | GO:0014028 | notochord formation(GO:0014028) |
| 0.4 | 1.1 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) positive regulation of aminoacyl-tRNA ligase activity(GO:1903632) regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.4 | 3.3 | GO:0085020 | protein K6-linked ubiquitination(GO:0085020) |
| 0.4 | 0.7 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.4 | 0.7 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.3 | 1.0 | GO:2000314 | negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.3 | 10.7 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.3 | 1.4 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.3 | 1.7 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.3 | 1.3 | GO:0046210 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.3 | 2.3 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.3 | 3.0 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.3 | 1.6 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.3 | 2.3 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.3 | 1.3 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.3 | 3.6 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.3 | 1.3 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.3 | 2.2 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.3 | 1.0 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.3 | 1.0 | GO:0060290 | transdifferentiation(GO:0060290) |
| 0.3 | 1.6 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.3 | 0.9 | GO:0061743 | motor learning(GO:0061743) |
| 0.3 | 2.2 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.3 | 0.6 | GO:1901228 | positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.3 | 3.7 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.3 | 0.9 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 0.3 | 0.9 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.3 | 3.9 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.3 | 3.9 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.3 | 0.3 | GO:0033122 | regulation of purine nucleotide catabolic process(GO:0033121) negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.3 | 4.4 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.3 | 2.0 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.3 | 3.1 | GO:1901250 | regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.3 | 3.1 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.3 | 0.8 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.3 | 1.4 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.3 | 0.8 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.3 | 5.5 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.3 | 2.7 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.3 | 1.6 | GO:1903755 | regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 0.3 | 1.3 | GO:0019659 | fermentation(GO:0006113) lactate biosynthetic process from pyruvate(GO:0019244) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.3 | 1.3 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.3 | 2.3 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.3 | 3.1 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.3 | 8.8 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.3 | 0.5 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.2 | 0.7 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.2 | 0.7 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.2 | 1.7 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.2 | 1.2 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.2 | 6.9 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.2 | 1.0 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.2 | 0.7 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.2 | 0.7 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.2 | 0.9 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.2 | 0.9 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.2 | 2.5 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.2 | 0.7 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.2 | 0.9 | GO:0001692 | histamine metabolic process(GO:0001692) histidine metabolic process(GO:0006547) imidazole-containing compound catabolic process(GO:0052805) |
| 0.2 | 0.9 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.2 | 2.0 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.2 | 2.0 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.2 | 0.9 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.2 | 1.3 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.2 | 2.4 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.2 | 1.3 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.2 | 3.7 | GO:0030220 | platelet formation(GO:0030220) |
| 0.2 | 1.1 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) convergent extension involved in gastrulation(GO:0060027) |
| 0.2 | 3.4 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.2 | 9.5 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.2 | 0.4 | GO:0010455 | positive regulation of cell fate commitment(GO:0010455) regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.2 | 1.2 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.2 | 0.4 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.2 | 0.4 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.2 | 1.4 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.2 | 0.8 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.2 | 1.6 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.2 | 1.0 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.2 | 1.2 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.2 | 2.1 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.2 | 3.5 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.2 | 0.2 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.2 | 0.8 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.2 | 3.9 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.2 | 1.1 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.2 | 0.8 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.2 | 2.6 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.2 | 8.3 | GO:0035307 | positive regulation of protein dephosphorylation(GO:0035307) |
| 0.2 | 0.6 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.2 | 0.4 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.2 | 0.6 | GO:0021972 | corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.2 | 0.7 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.2 | 1.5 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.2 | 5.8 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.2 | 0.5 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.2 | 0.5 | GO:1990859 | cellular response to endothelin(GO:1990859) |
| 0.2 | 3.8 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.2 | 0.9 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.2 | 1.9 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.2 | 0.8 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.2 | 0.7 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.2 | 1.2 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.2 | 0.7 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.2 | 0.7 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.2 | 3.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.2 | 0.5 | GO:0009838 | abscission(GO:0009838) |
| 0.2 | 0.7 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.2 | 0.8 | GO:0031424 | keratinization(GO:0031424) |
| 0.2 | 3.3 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.2 | 1.0 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.2 | 0.3 | GO:0071455 | cellular response to hyperoxia(GO:0071455) |
| 0.2 | 0.8 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.2 | 1.3 | GO:0072189 | ureter development(GO:0072189) |
| 0.2 | 1.7 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.2 | 1.3 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.2 | 1.4 | GO:0007512 | adult heart development(GO:0007512) |
| 0.2 | 1.1 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.2 | 0.8 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.2 | 3.4 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.2 | 0.9 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.2 | 1.7 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.2 | 0.5 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.2 | 1.1 | GO:0007320 | insemination(GO:0007320) |
| 0.2 | 0.6 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.2 | 0.9 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 10.8 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.2 | 0.5 | GO:0008050 | courtship behavior(GO:0007619) female courtship behavior(GO:0008050) |
| 0.2 | 1.7 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.1 | 2.2 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 0.6 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.1 | 0.7 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 5.0 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 3.8 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 0.9 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.1 | 2.8 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.6 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.1 | 2.5 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.1 | 0.4 | GO:0045056 | transcytosis(GO:0045056) |
| 0.1 | 1.6 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 2.9 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.1 | 0.4 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.1 | 1.0 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 1.4 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 0.4 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 | 4.2 | GO:1902808 | positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.1 | 0.4 | GO:0003192 | mitral valve formation(GO:0003192) cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.1 | 0.3 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) |
| 0.1 | 0.8 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 1.0 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 1.0 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.1 | 0.6 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 0.6 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 0.5 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.1 | 1.3 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.1 | 1.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.9 | GO:1901142 | insulin metabolic process(GO:1901142) |
| 0.1 | 0.4 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.1 | 1.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 2.9 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.1 | 0.9 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 2.8 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 3.1 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 5.4 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.1 | 0.7 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.1 | 1.4 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 1.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 2.3 | GO:0006073 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.1 | 0.7 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 1.5 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.1 | 1.2 | GO:0048733 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) sebaceous gland development(GO:0048733) |
| 0.1 | 0.4 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.1 | 0.4 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 4.1 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.1 | 1.8 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.1 | 2.0 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 0.6 | GO:0040031 | snRNA pseudouridine synthesis(GO:0031120) snRNA modification(GO:0040031) |
| 0.1 | 0.3 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.1 | 8.6 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.1 | 0.8 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.9 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 0.3 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) formin-nucleated actin cable assembly(GO:0070649) |
| 0.1 | 0.6 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.3 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.1 | 0.8 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.1 | 0.5 | GO:0061009 | common bile duct development(GO:0061009) |
| 0.1 | 0.5 | GO:0051189 | molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.1 | 1.4 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 0.3 | GO:0006244 | pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide metabolic process(GO:0009219) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.1 | 2.8 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 2.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.5 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 1.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 2.2 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.1 | 0.8 | GO:0021794 | thalamus development(GO:0021794) |
| 0.1 | 2.0 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 | 0.4 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.1 | 0.4 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.1 | 1.1 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.1 | 0.3 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.7 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.8 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.1 | 0.1 | GO:0006404 | RNA import into nucleus(GO:0006404) |
| 0.1 | 1.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 0.4 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.1 | 1.2 | GO:0045830 | positive regulation of isotype switching(GO:0045830) |
| 0.1 | 0.5 | GO:1904874 | positive regulation of telomerase RNA localization to Cajal body(GO:1904874) |
| 0.1 | 0.6 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 0.6 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 1.0 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.1 | 0.5 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.1 | 1.7 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.1 | 0.6 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.1 | 0.1 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.1 | 5.4 | GO:2000177 | regulation of neural precursor cell proliferation(GO:2000177) |
| 0.1 | 1.2 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 1.5 | GO:0010883 | regulation of lipid storage(GO:0010883) |
| 0.1 | 5.0 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 0.3 | GO:0048262 | determination of dorsal/ventral asymmetry(GO:0048262) determination of dorsal identity(GO:0048263) |
| 0.1 | 0.5 | GO:0021924 | cell proliferation in external granule layer(GO:0021924) cerebellar granule cell precursor proliferation(GO:0021930) |
| 0.1 | 1.7 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.5 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.1 | 0.8 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 | 0.2 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.7 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.1 | GO:0060672 | negative regulation of keratinocyte differentiation(GO:0045617) epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 3.3 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.1 | 0.2 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.1 | 0.9 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.1 | 0.6 | GO:0071459 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.1 | 0.2 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 1.3 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.1 | 0.2 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 0.1 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.1 | 0.1 | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.1 | 1.4 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 1.8 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.1 | 0.5 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 0.9 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.1 | 0.4 | GO:0045840 | positive regulation of mitotic nuclear division(GO:0045840) |
| 0.1 | 0.2 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.1 | 0.4 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 0.3 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 0.4 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.1 | 0.1 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.1 | 1.5 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.5 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.1 | 1.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.4 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.7 | GO:1904816 | regulation of protein localization to chromosome, telomeric region(GO:1904814) positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.1 | 0.9 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 0.2 | GO:0033580 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.1 | 0.3 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.1 | 0.3 | GO:0045794 | negative regulation of cell volume(GO:0045794) |
| 0.1 | 0.3 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.9 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 0.1 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.1 | 0.4 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 2.5 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 0.6 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.1 | 0.3 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.1 | 0.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.2 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 0.2 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.1 | 2.5 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 1.8 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.3 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.0 | 0.5 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.2 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.3 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.4 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.0 | 0.9 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 1.3 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.6 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.2 | GO:0060412 | ventricular septum morphogenesis(GO:0060412) |
| 0.0 | 1.0 | GO:0001825 | blastocyst formation(GO:0001825) |
| 0.0 | 0.4 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.7 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 3.5 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 1.2 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 1.4 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.5 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.1 | GO:0016132 | brassinosteroid metabolic process(GO:0016131) brassinosteroid biosynthetic process(GO:0016132) |
| 0.0 | 0.6 | GO:1903077 | negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.1 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) |
| 0.0 | 1.2 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.0 | 0.8 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.1 | GO:0001711 | endodermal cell fate commitment(GO:0001711) endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.6 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.2 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.0 | 1.4 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.4 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.6 | GO:0006739 | NADP metabolic process(GO:0006739) |
| 0.0 | 0.2 | GO:0060213 | positive regulation of mRNA 3'-end processing(GO:0031442) regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.2 | GO:0018195 | peptidyl-arginine modification(GO:0018195) |
| 0.0 | 0.2 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.2 | GO:0014037 | Schwann cell differentiation(GO:0014037) |
| 0.0 | 0.2 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.0 | 0.4 | GO:1901032 | negative regulation of response to reactive oxygen species(GO:1901032) negative regulation of hydrogen peroxide-induced cell death(GO:1903206) |
| 0.0 | 0.5 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.0 | 0.1 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.4 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.3 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 | 1.8 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.3 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.2 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.5 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 3.6 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.7 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 0.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 4.5 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.0 | 0.8 | GO:0017004 | cytochrome complex assembly(GO:0017004) |
| 0.0 | 0.8 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.0 | 0.7 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.2 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.0 | 0.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.3 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.1 | GO:0051352 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) |
| 0.0 | 2.0 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 0.2 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.4 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.0 | 1.1 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.5 | GO:0035411 | catenin import into nucleus(GO:0035411) |
| 0.0 | 0.1 | GO:0061038 | uterus morphogenesis(GO:0061038) |
| 0.0 | 0.3 | GO:0060760 | positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.0 | 0.7 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.3 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.0 | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) |
| 0.0 | 0.4 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 0.1 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.0 | 0.2 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.0 | 0.1 | GO:0033108 | mitochondrial respiratory chain complex assembly(GO:0033108) |
| 0.0 | 0.4 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.0 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.2 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.2 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.0 | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.0 | 0.2 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0010575 | positive regulation of vascular endothelial growth factor production(GO:0010575) |
| 0.0 | 0.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.8 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.3 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.2 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.0 | 0.6 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 7.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 1.4 | 5.6 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 1.4 | 11.0 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 1.4 | 4.1 | GO:0071914 | prominosome(GO:0071914) |
| 1.3 | 11.7 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.9 | 3.7 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.9 | 4.5 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.9 | 4.5 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.8 | 9.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.8 | 7.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.8 | 2.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.8 | 2.3 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.8 | 2.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.7 | 2.0 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.7 | 1.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.6 | 2.5 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 0.6 | 2.4 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.6 | 2.4 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.6 | 2.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.5 | 3.3 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.5 | 1.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.5 | 5.0 | GO:0005902 | microvillus(GO:0005902) |
| 0.4 | 2.2 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.4 | 2.5 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.4 | 1.2 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.4 | 3.5 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.4 | 2.0 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.4 | 2.3 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.4 | 27.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.4 | 4.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.4 | 5.9 | GO:0070938 | contractile ring(GO:0070938) |
| 0.4 | 1.5 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.4 | 4.7 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.4 | 3.6 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.3 | 0.3 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.3 | 2.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.3 | 1.3 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.3 | 1.3 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.3 | 0.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.3 | 4.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 0.6 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.3 | 2.1 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.3 | 3.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.3 | 0.9 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.3 | 3.9 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.3 | 0.3 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.3 | 3.7 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.3 | 0.9 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 0.3 | 1.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.3 | 2.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.3 | 5.3 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.3 | 4.7 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.3 | 1.4 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.3 | 2.4 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.3 | 1.6 | GO:1990356 | sumoylated E2 ligase complex(GO:1990356) |
| 0.3 | 14.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 1.0 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.2 | 4.4 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.2 | 3.1 | GO:0042581 | specific granule(GO:0042581) |
| 0.2 | 0.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 4.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 2.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 5.8 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 5.7 | GO:0001741 | XY body(GO:0001741) |
| 0.2 | 0.7 | GO:0000811 | GINS complex(GO:0000811) |
| 0.2 | 3.5 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.2 | 3.5 | GO:0051286 | cell tip(GO:0051286) |
| 0.2 | 0.6 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.2 | 3.0 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 0.8 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.2 | 3.8 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.2 | 2.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 0.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 1.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.2 | 0.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.2 | 1.3 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 0.9 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.2 | 0.7 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.2 | 0.9 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.2 | 2.5 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.2 | 2.8 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.2 | 0.9 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.2 | 1.7 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.2 | 1.0 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.2 | 1.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) replisome(GO:0030894) nuclear replisome(GO:0043601) |
| 0.2 | 0.8 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 2.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 1.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.2 | 7.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 0.5 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.2 | 1.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.2 | 0.5 | GO:0098536 | deuterosome(GO:0098536) |
| 0.2 | 0.5 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.2 | 1.4 | GO:0032982 | myosin filament(GO:0032982) |
| 0.2 | 0.5 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 1.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.7 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.1 | 2.2 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 1.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 1.9 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.3 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 2.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 1.4 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.1 | 1.9 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 1.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.4 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 0.9 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.6 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 1.5 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 3.5 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 1.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 1.6 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 0.7 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 5.4 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 1.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.2 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.1 | 3.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 1.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.8 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 1.0 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.6 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 2.7 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.9 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.5 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 1.0 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 1.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.9 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 0.8 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 3.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.2 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.1 | 5.3 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 13.1 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.1 | 1.0 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.4 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 8.6 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 4.1 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 2.9 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 1.0 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 0.6 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 5.2 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.1 | 0.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.6 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 0.6 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 0.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 8.3 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 0.5 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 0.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 2.1 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.4 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 1.2 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 0.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 2.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 1.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 3.4 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.1 | 10.8 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 0.4 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 3.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.3 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.9 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.6 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.3 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 1.5 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 2.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.6 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 1.9 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 1.8 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 2.2 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.3 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.5 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.9 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 1.2 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 1.5 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 1.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 1.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.3 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.8 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.8 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.4 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.4 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 1.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.5 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 0.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.2 | GO:0005832 | zona pellucida receptor complex(GO:0002199) chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 1.8 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.1 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.5 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 15.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 2.6 | 7.9 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 2.6 | 7.7 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 2.1 | 6.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 1.4 | 9.6 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 1.4 | 6.8 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 1.1 | 10.6 | GO:0019956 | chemokine binding(GO:0019956) |
| 1.0 | 4.2 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 1.0 | 6.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 1.0 | 3.1 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 1.0 | 3.0 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 1.0 | 7.0 | GO:0070061 | fructose binding(GO:0070061) |
| 1.0 | 3.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.9 | 3.7 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.9 | 3.6 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.9 | 4.5 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.9 | 3.5 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.8 | 5.0 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.8 | 4.0 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.8 | 2.3 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.7 | 8.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.7 | 5.9 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.7 | 5.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.7 | 5.4 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.6 | 2.5 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.6 | 2.5 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.6 | 5.0 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.6 | 4.3 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.6 | 2.4 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.6 | 3.0 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.6 | 2.4 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.6 | 2.3 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.6 | 2.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.6 | 2.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.6 | 2.8 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.5 | 1.6 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.5 | 1.1 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.5 | 2.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.5 | 1.6 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.5 | 1.6 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.5 | 4.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.5 | 2.0 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.5 | 1.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.5 | 3.4 | GO:1901612 | phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 0.5 | 2.4 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.5 | 1.9 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.5 | 2.4 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.5 | 3.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.5 | 4.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.4 | 0.4 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.4 | 2.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.4 | 9.6 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.4 | 2.5 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.4 | 4.6 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.4 | 9.8 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.4 | 1.2 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.4 | 2.0 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.4 | 1.9 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.4 | 2.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.4 | 6.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.4 | 1.5 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.4 | 1.5 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.4 | 1.8 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.4 | 11.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.4 | 1.4 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.4 | 0.7 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.4 | 0.4 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.3 | 3.5 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.3 | 1.4 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.3 | 2.4 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.3 | 6.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.3 | 1.3 | GO:0016708 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.3 | 1.7 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.3 | 2.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.3 | 2.0 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.3 | 1.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.3 | 1.3 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.3 | 3.5 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.3 | 10.4 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.3 | 0.9 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.3 | 0.9 | GO:0046592 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.3 | 0.9 | GO:0018738 | S-formylglutathione hydrolase activity(GO:0018738) |
| 0.3 | 2.3 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.3 | 2.6 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.3 | 1.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.3 | 2.0 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.3 | 0.8 | GO:0001565 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.3 | 8.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.3 | 1.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.3 | 1.6 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.3 | 2.6 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.3 | 0.8 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.3 | 1.3 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.2 | 0.7 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) U7 snRNA binding(GO:0071209) |
| 0.2 | 3.9 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 11.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 1.2 | GO:0070404 | NADH binding(GO:0070404) |
| 0.2 | 2.7 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.2 | 0.7 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.2 | 1.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 0.9 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.2 | 0.9 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.2 | 1.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.2 | 5.7 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.2 | 2.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.2 | 1.5 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.2 | 1.3 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.2 | 3.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 0.2 | GO:0032405 | MutLalpha complex binding(GO:0032405) |
| 0.2 | 1.6 | GO:0046790 | virion binding(GO:0046790) |
| 0.2 | 0.8 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.2 | 2.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.2 | 1.4 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.2 | 1.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 1.4 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.2 | 1.8 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.2 | 0.8 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.2 | 5.0 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 1.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.2 | 6.5 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.2 | 1.8 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.2 | 1.0 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 5.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 0.6 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.2 | 3.5 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.2 | 0.7 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.2 | 0.7 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 6.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 2.7 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.2 | 1.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 1.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.2 | 1.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 1.0 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.2 | 7.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.2 | 1.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.2 | 0.7 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.2 | 2.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.2 | 0.5 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.2 | 0.5 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.2 | 0.6 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.2 | 1.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 1.7 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.2 | 0.6 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.2 | 4.2 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.2 | 1.8 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.2 | 1.2 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.2 | 0.5 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.2 | 0.9 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 2.8 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.7 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 0.9 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 1.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 1.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.8 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.9 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 0.7 | GO:0044020 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.1 | 0.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 5.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 2.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 3.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 1.8 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.1 | 0.5 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.1 | 4.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.0 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 20.1 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 0.5 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 6.1 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.7 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.5 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.1 | 1.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 0.6 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.3 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 0.3 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.1 | 9.1 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 15.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 0.4 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 5.4 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 0.9 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 1.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 0.4 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.4 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 0.2 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 1.0 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 1.0 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 2.6 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 2.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.9 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.9 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.1 | 7.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 0.6 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.7 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 0.4 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 0.4 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 2.9 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.1 | 0.8 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.1 | 0.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 0.6 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.1 | 0.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 1.9 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 5.0 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 0.4 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 1.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.6 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.1 | 0.3 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.1 | 0.5 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 0.4 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 1.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 3.5 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.1 | 1.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.6 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 1.0 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 1.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.5 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.5 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.4 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.1 | 0.5 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.1 | 9.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 1.9 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 1.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.8 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 1.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.5 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 0.8 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 3.9 | GO:0008186 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.1 | 2.2 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 1.0 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 2.7 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.1 | 1.8 | GO:0051287 | NAD binding(GO:0051287) |
| 0.1 | 14.5 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.4 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.8 | GO:0030553 | cGMP binding(GO:0030553) 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 1.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.9 | GO:0015144 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.0 | 0.5 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.2 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.3 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.2 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 1.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 0.2 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.2 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.0 | 0.4 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.7 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 1.4 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.2 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.2 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.0 | 0.2 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.0 | 0.2 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0009918 | sterol delta7 reductase activity(GO:0009918) 7-dehydrocholesterol reductase activity(GO:0047598) |
| 0.0 | 0.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.2 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.6 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 3.6 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.9 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.3 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.1 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 1.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.8 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.0 | 0.2 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.5 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.3 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.2 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 1.3 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.3 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.7 | GO:0035326 | enhancer binding(GO:0035326) |
| 0.0 | 0.3 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.2 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 1.2 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 0.2 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.6 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.6 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.3 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
| 0.0 | 0.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.0 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 1.8 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 0.6 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 1.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.1 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 11.6 | GO:0003677 | DNA binding(GO:0003677) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 7.3 | ST_TYPE_I_INTERFERON_PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.4 | 5.8 | PID_INTEGRIN2_PATHWAY | Beta2 integrin cell surface interactions |
| 0.4 | 12.4 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.3 | 4.5 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.3 | 18.9 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.3 | 0.5 | PID_IL3_PATHWAY | IL3-mediated signaling events |
| 0.3 | 13.1 | PID_FANCONI_PATHWAY | Fanconi anemia pathway |
| 0.3 | 4.8 | PID_INTEGRIN_CS_PATHWAY | Integrin family cell surface interactions |
| 0.2 | 8.5 | PID_AURORA_A_PATHWAY | Aurora A signaling |
| 0.2 | 1.1 | PID_CD8_TCR_DOWNSTREAM_PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.2 | 8.5 | PID_ARF6_PATHWAY | Arf6 signaling events |
| 0.2 | 8.3 | PID_RXR_VDR_PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 9.5 | PID_HES_HEY_PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 1.3 | PID_INTEGRIN4_PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 12.2 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 3.4 | PID_UPA_UPAR_PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 2.8 | PID_BARD1_PATHWAY | BARD1 signaling events |
| 0.2 | 5.7 | PID_P38_MK2_PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 17.8 | PID_SMAD2_3NUCLEAR_PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.2 | 3.4 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 3.3 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.1 | 3.5 | PID_PDGFRA_PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 0.9 | PID_A6B1_A6B4_INTEGRIN_PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 2.1 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 2.9 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.1 | 0.7 | PID_RETINOIC_ACID_PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 1.4 | PID_RANBP2_PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 0.8 | PID_EPHA2_FWD_PATHWAY | EPHA2 forward signaling |
| 0.1 | 2.6 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.1 | 4.7 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 3.2 | PID_MYC_REPRESS_PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 3.3 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 3.5 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.1 | 0.7 | PID_IL6_7_PATHWAY | IL6-mediated signaling events |
| 0.1 | 3.3 | PID_FGF_PATHWAY | FGF signaling pathway |
| 0.1 | 1.4 | PID_INSULIN_GLUCOSE_PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 0.7 | PID_INTEGRIN_A9B1_PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 3.0 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 2.4 | PID_TRKR_PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 0.7 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.1 | 2.5 | PID_NOTCH_PATHWAY | Notch signaling pathway |
| 0.1 | 0.9 | PID_HIF1A_PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 3.3 | WNT_SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.7 | PID_ARF6_DOWNSTREAM_PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.5 | PID_NFAT_TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.2 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID_ERB_GENOMIC_PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.6 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.5 | PID_CIRCADIAN_PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.4 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.0 | 0.7 | PID_P38_MKK3_6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.8 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.1 | PID_ECADHERIN_STABILIZATION_PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.5 | PID_SHP2_PATHWAY | SHP2 signaling |
| 0.0 | 0.6 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.0 | 0.2 | PID_ANTHRAX_PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 2.3 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.0 | 0.2 | PID_RHODOPSIN_PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.6 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 7.2 | REACTOME_SYNTHESIS_OF_BILE_ACIDS_AND_BILE_SALTS_VIA_24_HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.7 | 11.4 | REACTOME_ASSOCIATION_OF_LICENSING_FACTORS_WITH_THE_PRE_REPLICATIVE_COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.7 | 17.8 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.6 | 9.6 | REACTOME_PROCESSIVE_SYNTHESIS_ON_THE_LAGGING_STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.6 | 11.6 | REACTOME_RNA_POL_I_PROMOTER_OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.5 | 2.2 | REACTOME_ETHANOL_OXIDATION | Genes involved in Ethanol oxidation |
| 0.5 | 7.7 | REACTOME_APOPTOSIS_INDUCED_DNA_FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.4 | 14.4 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.4 | 23.9 | REACTOME_DEPOSITION_OF_NEW_CENPA_CONTAINING_NUCLEOSOMES_AT_THE_CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.3 | 7.8 | REACTOME_CIRCADIAN_REPRESSION_OF_EXPRESSION_BY_REV_ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.3 | 6.7 | REACTOME_FANCONI_ANEMIA_PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.3 | 6.9 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.3 | 6.6 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.3 | 2.5 | REACTOME_HYALURONAN_UPTAKE_AND_DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.3 | 4.3 | REACTOME_MITOCHONDRIAL_FATTY_ACID_BETA_OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.2 | 5.6 | REACTOME_GABA_SYNTHESIS_RELEASE_REUPTAKE_AND_DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 6.1 | REACTOME_PRE_NOTCH_TRANSCRIPTION_AND_TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.2 | 9.1 | REACTOME_GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 0.7 | REACTOME_DNA_STRAND_ELONGATION | Genes involved in DNA strand elongation |
| 0.2 | 2.8 | REACTOME_SIGNALING_BY_FGFR | Genes involved in Signaling by FGFR |
| 0.2 | 1.8 | REACTOME_TRYPTOPHAN_CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 17.8 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 3.1 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.2 | 1.9 | REACTOME_PROTEOLYTIC_CLEAVAGE_OF_SNARE_COMPLEX_PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 2.7 | REACTOME_PURINE_RIBONUCLEOSIDE_MONOPHOSPHATE_BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 2.4 | REACTOME_CHYLOMICRON_MEDIATED_LIPID_TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 1.0 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELL_ADHESION_PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 1.9 | REACTOME_GAP_JUNCTION_ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 1.6 | REACTOME_G1_S_SPECIFIC_TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.2 | 2.0 | REACTOME_RECYCLING_PATHWAY_OF_L1 | Genes involved in Recycling pathway of L1 |
| 0.2 | 1.4 | REACTOME_FORMATION_OF_INCISION_COMPLEX_IN_GG_NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 2.4 | REACTOME_GLYCOGEN_BREAKDOWN_GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 2.4 | REACTOME_BMAL1_CLOCK_NPAS2_ACTIVATES_CIRCADIAN_EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 1.6 | REACTOME_SIGNALING_BY_NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.2 | REACTOME_DESTABILIZATION_OF_MRNA_BY_BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 1.6 | REACTOME_SLBP_DEPENDENT_PROCESSING_OF_REPLICATION_DEPENDENT_HISTONE_PRE_MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 2.3 | REACTOME_VIRAL_MESSENGER_RNA_SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 2.2 | REACTOME_PRE_NOTCH_PROCESSING_IN_GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 3.9 | REACTOME_ENOS_ACTIVATION_AND_REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 3.8 | REACTOME_RNA_POL_II_TRANSCRIPTION_PRE_INITIATION_AND_PROMOTER_OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 1.3 | REACTOME_FGFR2C_LIGAND_BINDING_AND_ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.1 | 7.8 | REACTOME_APC_C_CDC20_MEDIATED_DEGRADATION_OF_MITOTIC_PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.1 | 2.8 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.0 | REACTOME_REGULATION_OF_KIT_SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 6.3 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.1 | 2.9 | REACTOME_ACTIVATION_OF_ATR_IN_RESPONSE_TO_REPLICATION_STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.1 | 3.4 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.9 | REACTOME_GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.7 | REACTOME_FORMATION_OF_ATP_BY_CHEMIOSMOTIC_COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 4.2 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELLULAR_PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 6.7 | REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 8.9 | REACTOME_PEPTIDE_CHAIN_ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 1.5 | REACTOME_TRAF6_MEDIATED_IRF7_ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 0.5 | REACTOME_REGULATION_OF_ORNITHINE_DECARBOXYLASE_ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 2.6 | REACTOME_GLUCOSE_TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 0.6 | REACTOME_REGULATION_OF_IFNG_SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 2.8 | REACTOME_ANTIVIRAL_MECHANISM_BY_IFN_STIMULATED_GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 1.4 | REACTOME_PYRUVATE_METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 1.6 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.5 | REACTOME_CITRIC_ACID_CYCLE_TCA_CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 0.4 | REACTOME_PROLACTIN_RECEPTOR_SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 2.7 | REACTOME_NETRIN1_SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 0.9 | REACTOME_METABOLISM_OF_POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.3 | REACTOME_NEPHRIN_INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 0.7 | REACTOME_BASE_EXCISION_REPAIR | Genes involved in Base Excision Repair |
| 0.1 | 0.3 | REACTOME_REGULATION_OF_COMPLEMENT_CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.2 | REACTOME_ACTIVATED_AMPK_STIMULATES_FATTY_ACID_OXIDATION_IN_MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 1.7 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.7 | REACTOME_INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.3 | REACTOME_SYNTHESIS_OF_BILE_ACIDS_AND_BILE_SALTS_VIA_7ALPHA_HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.8 | REACTOME_MUSCLE_CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 1.1 | REACTOME_REGULATION_OF_BETA_CELL_DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.9 | REACTOME_AMINO_ACID_SYNTHESIS_AND_INTERCONVERSION_TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.8 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 4.5 | REACTOME_MRNA_SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.7 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.8 | REACTOME_CGMP_EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.7 | REACTOME_3_UTR_MEDIATED_TRANSLATIONAL_REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.0 | 0.8 | REACTOME_MITOCHONDRIAL_TRNA_AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME_PYRIMIDINE_CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.8 | REACTOME_MEIOTIC_SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.2 | REACTOME_TRAF6_MEDIATED_IRF7_ACTIVATION_IN_TLR7_8_OR_9_SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 1.8 | REACTOME_TRANSCRIPTIONAL_REGULATION_OF_WHITE_ADIPOCYTE_DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.2 | REACTOME_OLFACTORY_SIGNALING_PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.4 | REACTOME_METABOLISM_OF_NON_CODING_RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.9 | REACTOME_SYNTHESIS_OF_PA | Genes involved in Synthesis of PA |
| 0.0 | 1.5 | REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.8 | REACTOME_POST_TRANSLATIONAL_MODIFICATION_SYNTHESIS_OF_GPI_ANCHORED_PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.1 | REACTOME_SEMA3A_PAK_DEPENDENT_AXON_REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.4 | REACTOME_METABOLISM_OF_PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME_MRNA_DECAY_BY_5_TO_3_EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.3 | REACTOME_GENERIC_TRANSCRIPTION_PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.0 | REACTOME_GPVI_MEDIATED_ACTIVATION_CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.2 | REACTOME_MEIOTIC_RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.6 | REACTOME_TRANSLATION | Genes involved in Translation |
| 0.0 | 0.7 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | Genes involved in Respiratory electron transport |