Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
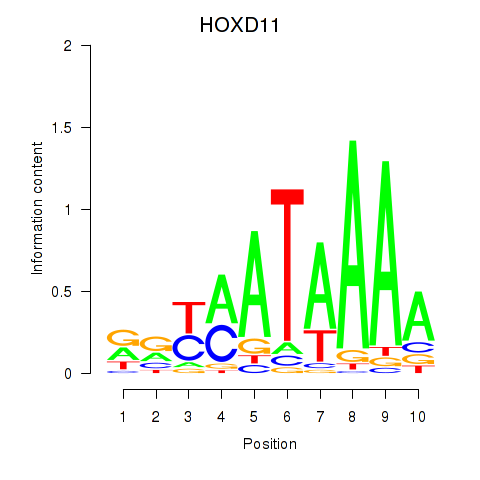
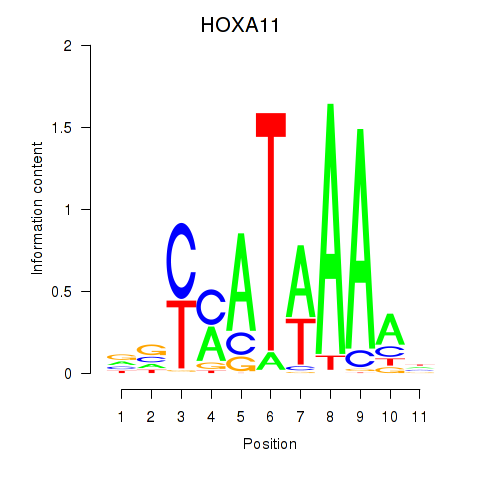
Results for HOXD11_HOXA11
Z-value: 0.44
Transcription factors associated with HOXD11_HOXA11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD11
|
ENSG00000128713.11 | homeobox D11 |
|
HOXA11
|
ENSG00000005073.5 | homeobox A11 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA11 | hg19_v2_chr7_-_27224795_27224840 | -0.59 | 2.2e-01 | Click! |
Activity profile of HOXD11_HOXA11 motif
Sorted Z-values of HOXD11_HOXA11 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_35658007 | 0.36 |
ENST00000602361.1
|
RMRP
|
RNA component of mitochondrial RNA processing endoribonuclease |
| chr5_-_159766528 | 0.31 |
ENST00000505287.2
|
CCNJL
|
cyclin J-like |
| chr2_+_38177575 | 0.25 |
ENST00000407257.1
ENST00000417700.2 ENST00000234195.3 ENST00000442857.1 |
RMDN2
|
regulator of microtubule dynamics 2 |
| chr14_-_60952739 | 0.23 |
ENST00000555476.1
ENST00000321731.3 |
C14orf39
|
chromosome 14 open reading frame 39 |
| chr8_-_71157595 | 0.22 |
ENST00000519724.1
|
NCOA2
|
nuclear receptor coactivator 2 |
| chr2_+_181988620 | 0.19 |
ENST00000428474.1
ENST00000424655.1 |
AC104820.2
|
AC104820.2 |
| chr13_+_113030658 | 0.19 |
ENST00000414180.1
ENST00000443541.1 |
SPACA7
|
sperm acrosome associated 7 |
| chr1_+_192127578 | 0.17 |
ENST00000367460.3
|
RGS18
|
regulator of G-protein signaling 18 |
| chr6_-_26189304 | 0.17 |
ENST00000340756.2
|
HIST1H4D
|
histone cluster 1, H4d |
| chr1_-_168464875 | 0.17 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr1_-_230991747 | 0.17 |
ENST00000523410.1
|
C1orf198
|
chromosome 1 open reading frame 198 |
| chr14_+_73563735 | 0.16 |
ENST00000532192.1
|
RBM25
|
RNA binding motif protein 25 |
| chr12_-_94673956 | 0.16 |
ENST00000551941.1
|
RP11-1105G2.3
|
Uncharacterized protein |
| chr19_-_33182616 | 0.16 |
ENST00000592431.1
|
CTD-2538C1.2
|
CTD-2538C1.2 |
| chr14_+_39944025 | 0.15 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr12_-_71148413 | 0.14 |
ENST00000440835.2
ENST00000549308.1 ENST00000550661.1 |
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr10_+_85933494 | 0.14 |
ENST00000372126.3
|
C10orf99
|
chromosome 10 open reading frame 99 |
| chr12_+_34175398 | 0.14 |
ENST00000538927.1
|
ALG10
|
ALG10, alpha-1,2-glucosyltransferase |
| chr6_+_71104588 | 0.14 |
ENST00000418403.1
|
RP11-462G2.1
|
RP11-462G2.1 |
| chr7_+_79763271 | 0.13 |
ENST00000442586.1
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr6_-_15548591 | 0.13 |
ENST00000509674.1
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr8_-_95220775 | 0.13 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr7_+_138915102 | 0.12 |
ENST00000486663.1
|
UBN2
|
ubinuclein 2 |
| chrX_+_123097014 | 0.12 |
ENST00000394478.1
|
STAG2
|
stromal antigen 2 |
| chr20_+_58713514 | 0.12 |
ENST00000432910.1
|
RP5-1043L13.1
|
RP5-1043L13.1 |
| chr20_-_45530365 | 0.12 |
ENST00000414085.1
|
RP11-323C15.2
|
RP11-323C15.2 |
| chr3_+_186383741 | 0.12 |
ENST00000232003.4
|
HRG
|
histidine-rich glycoprotein |
| chr12_+_6833323 | 0.11 |
ENST00000544725.1
|
COPS7A
|
COP9 signalosome subunit 7A |
| chr2_-_160568529 | 0.11 |
ENST00000418770.1
|
AC009961.3
|
AC009961.3 |
| chr14_+_35514323 | 0.11 |
ENST00000555211.1
|
FAM177A1
|
family with sequence similarity 177, member A1 |
| chr2_-_3504587 | 0.11 |
ENST00000415131.1
|
ADI1
|
acireductone dioxygenase 1 |
| chr1_+_120254510 | 0.10 |
ENST00000369409.4
|
PHGDH
|
phosphoglycerate dehydrogenase |
| chr16_-_21663950 | 0.10 |
ENST00000268389.4
|
IGSF6
|
immunoglobulin superfamily, member 6 |
| chr19_-_54618650 | 0.10 |
ENST00000391757.1
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr2_+_109204909 | 0.10 |
ENST00000393310.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr9_+_109685630 | 0.10 |
ENST00000451160.2
|
RP11-508N12.4
|
Uncharacterized protein |
| chr6_+_12958137 | 0.10 |
ENST00000457702.2
ENST00000379345.2 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr4_-_185694872 | 0.10 |
ENST00000505492.1
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1 |
| chr2_-_207078086 | 0.09 |
ENST00000442134.1
|
GPR1
|
G protein-coupled receptor 1 |
| chr2_+_220306238 | 0.09 |
ENST00000435853.1
|
SPEG
|
SPEG complex locus |
| chr8_+_125486939 | 0.09 |
ENST00000303545.3
|
RNF139
|
ring finger protein 139 |
| chr8_+_121925301 | 0.09 |
ENST00000517739.1
|
RP11-369K17.1
|
RP11-369K17.1 |
| chr17_+_58018269 | 0.09 |
ENST00000591035.1
|
RP11-178C3.1
|
Uncharacterized protein |
| chr17_+_75369400 | 0.09 |
ENST00000590059.1
|
SEPT9
|
septin 9 |
| chr7_+_73106926 | 0.09 |
ENST00000453316.1
|
WBSCR22
|
Williams Beuren syndrome chromosome region 22 |
| chr11_-_111649074 | 0.09 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr6_+_46761118 | 0.09 |
ENST00000230588.4
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase) |
| chr17_+_42785976 | 0.09 |
ENST00000393547.2
ENST00000398338.3 |
DBF4B
|
DBF4 homolog B (S. cerevisiae) |
| chr17_-_27188984 | 0.09 |
ENST00000582320.2
|
MIR144
|
microRNA 451b |
| chr13_-_47471155 | 0.08 |
ENST00000543956.1
ENST00000542664.1 |
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled |
| chr8_-_116681686 | 0.08 |
ENST00000519815.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr11_+_3968573 | 0.08 |
ENST00000532990.1
|
STIM1
|
stromal interaction molecule 1 |
| chr3_-_156534754 | 0.08 |
ENST00000472943.1
ENST00000473352.1 |
LINC00886
|
long intergenic non-protein coding RNA 886 |
| chr1_+_84630574 | 0.08 |
ENST00000413538.1
ENST00000417530.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr1_-_161207986 | 0.08 |
ENST00000506209.1
ENST00000367980.2 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr12_+_70219052 | 0.08 |
ENST00000552032.2
ENST00000547771.2 |
MYRFL
|
myelin regulatory factor-like |
| chr12_-_71148357 | 0.08 |
ENST00000378778.1
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr2_+_149974684 | 0.08 |
ENST00000450639.1
|
LYPD6B
|
LY6/PLAUR domain containing 6B |
| chr12_+_38710555 | 0.08 |
ENST00000551464.1
|
ALG10B
|
ALG10B, alpha-1,2-glucosyltransferase |
| chrX_+_37865804 | 0.08 |
ENST00000297875.2
ENST00000357972.5 |
SYTL5
|
synaptotagmin-like 5 |
| chr2_-_166702601 | 0.08 |
ENST00000428888.1
|
AC009495.4
|
AC009495.4 |
| chr6_+_12007963 | 0.08 |
ENST00000607445.1
|
RP11-456H18.2
|
RP11-456H18.2 |
| chr1_+_153004800 | 0.08 |
ENST00000392661.3
|
SPRR1B
|
small proline-rich protein 1B |
| chr1_+_92545862 | 0.08 |
ENST00000370382.3
ENST00000342818.3 |
BTBD8
|
BTB (POZ) domain containing 8 |
| chr12_-_6580094 | 0.08 |
ENST00000361716.3
|
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr9_+_67977438 | 0.08 |
ENST00000456982.1
|
RP11-195B21.3
|
Protein LOC644249 |
| chr18_+_21572737 | 0.08 |
ENST00000304621.6
|
TTC39C
|
tetratricopeptide repeat domain 39C |
| chr9_-_133814527 | 0.08 |
ENST00000451466.1
|
FIBCD1
|
fibrinogen C domain containing 1 |
| chr11_-_77791156 | 0.08 |
ENST00000281031.4
|
NDUFC2
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2, 14.5kDa |
| chr17_-_55911970 | 0.08 |
ENST00000581805.1
ENST00000580960.1 |
RP11-60A24.3
|
RP11-60A24.3 |
| chr9_+_125133467 | 0.07 |
ENST00000426608.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr11_+_85566422 | 0.07 |
ENST00000342404.3
|
CCDC83
|
coiled-coil domain containing 83 |
| chr19_-_10446449 | 0.07 |
ENST00000592439.1
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr1_-_152386732 | 0.07 |
ENST00000271835.3
|
CRNN
|
cornulin |
| chr12_-_76377795 | 0.07 |
ENST00000552856.1
|
RP11-114H23.1
|
RP11-114H23.1 |
| chr15_-_99789736 | 0.07 |
ENST00000560235.1
ENST00000394132.2 ENST00000560860.1 ENST00000558078.1 ENST00000394136.1 ENST00000262074.4 ENST00000558613.1 ENST00000394130.1 ENST00000560772.1 |
TTC23
|
tetratricopeptide repeat domain 23 |
| chr2_-_231989808 | 0.07 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr15_+_66797627 | 0.07 |
ENST00000565627.1
ENST00000564179.1 |
ZWILCH
|
zwilch kinetochore protein |
| chr15_+_36887069 | 0.07 |
ENST00000566807.1
ENST00000567389.1 ENST00000562877.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr3_+_140981456 | 0.07 |
ENST00000504264.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr4_-_76957214 | 0.07 |
ENST00000306621.3
|
CXCL11
|
chemokine (C-X-C motif) ligand 11 |
| chr1_+_28527070 | 0.07 |
ENST00000596102.1
|
AL353354.2
|
AL353354.2 |
| chr3_+_177159695 | 0.07 |
ENST00000442937.1
|
LINC00578
|
long intergenic non-protein coding RNA 578 |
| chr10_-_90712520 | 0.07 |
ENST00000224784.6
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr17_-_7760457 | 0.07 |
ENST00000576384.1
|
LSMD1
|
LSM domain containing 1 |
| chr12_-_6579808 | 0.06 |
ENST00000535180.1
ENST00000400911.3 |
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr4_+_169013666 | 0.06 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr14_-_20774092 | 0.06 |
ENST00000423949.2
ENST00000553828.1 ENST00000258821.3 |
TTC5
|
tetratricopeptide repeat domain 5 |
| chr7_+_128784712 | 0.06 |
ENST00000289407.4
|
TSPAN33
|
tetraspanin 33 |
| chr8_+_107738343 | 0.06 |
ENST00000521592.1
|
OXR1
|
oxidation resistance 1 |
| chr17_+_7211656 | 0.06 |
ENST00000416016.2
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr11_+_34664014 | 0.06 |
ENST00000527935.1
|
EHF
|
ets homologous factor |
| chr16_+_57653625 | 0.06 |
ENST00000567553.1
ENST00000565314.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr18_+_32556892 | 0.06 |
ENST00000591734.1
ENST00000413393.1 ENST00000589180.1 ENST00000587359.1 |
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr2_-_157198860 | 0.06 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr10_-_96829246 | 0.06 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr11_+_63606373 | 0.06 |
ENST00000402010.2
ENST00000315032.8 ENST00000377809.4 ENST00000413835.2 ENST00000377810.3 |
MARK2
|
MAP/microtubule affinity-regulating kinase 2 |
| chr11_-_796197 | 0.06 |
ENST00000530360.1
ENST00000528606.1 ENST00000320230.5 |
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr10_-_98031265 | 0.06 |
ENST00000224337.5
ENST00000371176.2 |
BLNK
|
B-cell linker |
| chr9_+_95736758 | 0.06 |
ENST00000337352.6
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr6_+_131571535 | 0.06 |
ENST00000474850.2
|
AKAP7
|
A kinase (PRKA) anchor protein 7 |
| chr17_-_38545799 | 0.06 |
ENST00000577541.1
|
TOP2A
|
topoisomerase (DNA) II alpha 170kDa |
| chr15_-_85197501 | 0.06 |
ENST00000434634.2
|
WDR73
|
WD repeat domain 73 |
| chr1_-_101360331 | 0.06 |
ENST00000416479.1
ENST00000370113.3 |
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr3_-_18480260 | 0.06 |
ENST00000454909.2
|
SATB1
|
SATB homeobox 1 |
| chr12_-_118796910 | 0.06 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr6_+_131958436 | 0.06 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr19_-_10420459 | 0.06 |
ENST00000403352.1
ENST00000403903.3 |
ZGLP1
|
zinc finger, GATA-like protein 1 |
| chrX_+_109602039 | 0.06 |
ENST00000520821.1
|
RGAG1
|
retrotransposon gag domain containing 1 |
| chrX_-_11129229 | 0.06 |
ENST00000608176.1
ENST00000433747.2 ENST00000608576.1 ENST00000608916.1 |
RP11-120D5.1
|
RP11-120D5.1 |
| chr4_+_187148556 | 0.06 |
ENST00000264690.6
ENST00000446598.2 ENST00000414291.1 ENST00000513864.1 |
KLKB1
|
kallikrein B, plasma (Fletcher factor) 1 |
| chr12_-_100486668 | 0.06 |
ENST00000550544.1
ENST00000551980.1 ENST00000548045.1 ENST00000545232.2 ENST00000551973.1 |
UHRF1BP1L
|
UHRF1 binding protein 1-like |
| chr1_-_242162375 | 0.06 |
ENST00000357246.3
|
MAP1LC3C
|
microtubule-associated protein 1 light chain 3 gamma |
| chr12_-_52828147 | 0.06 |
ENST00000252245.5
|
KRT75
|
keratin 75 |
| chr8_+_76452097 | 0.05 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr4_-_69434245 | 0.05 |
ENST00000317746.2
|
UGT2B17
|
UDP glucuronosyltransferase 2 family, polypeptide B17 |
| chr12_+_6833237 | 0.05 |
ENST00000229251.3
ENST00000539735.1 ENST00000538410.1 |
COPS7A
|
COP9 signalosome subunit 7A |
| chr2_+_109204743 | 0.05 |
ENST00000332345.6
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr3_-_108672742 | 0.05 |
ENST00000261047.3
|
GUCA1C
|
guanylate cyclase activator 1C |
| chr22_+_30821732 | 0.05 |
ENST00000355143.4
|
MTFP1
|
mitochondrial fission process 1 |
| chr8_+_52730143 | 0.05 |
ENST00000415643.1
|
AC090186.1
|
Uncharacterized protein |
| chr18_+_48918368 | 0.05 |
ENST00000583982.1
ENST00000578152.1 ENST00000583609.1 ENST00000435144.1 ENST00000580841.1 |
RP11-267C16.1
|
RP11-267C16.1 |
| chr3_+_167582561 | 0.05 |
ENST00000463642.1
ENST00000464514.1 |
RP11-298O21.6
RP11-298O21.7
|
RP11-298O21.6 RP11-298O21.7 |
| chr22_+_50981079 | 0.05 |
ENST00000609268.1
|
CTA-384D8.34
|
CTA-384D8.34 |
| chr13_-_46626820 | 0.05 |
ENST00000428921.1
|
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr4_+_144354644 | 0.05 |
ENST00000512843.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr12_+_43086018 | 0.05 |
ENST00000550177.1
|
RP11-25I15.3
|
RP11-25I15.3 |
| chr2_+_97454321 | 0.05 |
ENST00000540067.1
|
CNNM4
|
cyclin M4 |
| chr1_-_219615984 | 0.05 |
ENST00000420762.1
|
RP11-95P13.1
|
RP11-95P13.1 |
| chr4_-_84035905 | 0.05 |
ENST00000311507.4
|
PLAC8
|
placenta-specific 8 |
| chr5_+_59783941 | 0.05 |
ENST00000506884.1
ENST00000504876.2 |
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr1_+_65730385 | 0.05 |
ENST00000263441.7
ENST00000395325.3 |
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr11_+_34999328 | 0.05 |
ENST00000526309.1
|
PDHX
|
pyruvate dehydrogenase complex, component X |
| chr15_-_55488817 | 0.05 |
ENST00000569386.1
|
RSL24D1
|
ribosomal L24 domain containing 1 |
| chr19_+_19626531 | 0.05 |
ENST00000507754.4
|
NDUFA13
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13 |
| chr7_-_130080977 | 0.05 |
ENST00000223208.5
|
CEP41
|
centrosomal protein 41kDa |
| chr11_-_26593677 | 0.05 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chrX_+_65384052 | 0.05 |
ENST00000336279.5
ENST00000458621.1 |
HEPH
|
hephaestin |
| chr2_-_176046391 | 0.05 |
ENST00000392541.3
ENST00000409194.1 |
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr1_-_182369751 | 0.05 |
ENST00000367565.1
|
TEDDM1
|
transmembrane epididymal protein 1 |
| chr1_-_101360205 | 0.05 |
ENST00000450240.1
|
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr4_+_76439095 | 0.05 |
ENST00000506261.1
|
THAP6
|
THAP domain containing 6 |
| chr2_-_190446738 | 0.05 |
ENST00000427419.1
ENST00000455320.1 |
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr12_+_25205155 | 0.05 |
ENST00000550945.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr8_-_133772794 | 0.05 |
ENST00000519187.1
ENST00000523829.1 ENST00000356838.3 ENST00000377901.4 ENST00000519304.1 |
TMEM71
|
transmembrane protein 71 |
| chr1_-_101360374 | 0.05 |
ENST00000535414.1
|
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr19_-_5903714 | 0.05 |
ENST00000586349.1
ENST00000585661.1 ENST00000308961.4 ENST00000592634.1 ENST00000418389.2 ENST00000252675.5 |
AC024592.12
NDUFA11
FUT5
|
Uncharacterized protein NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa fucosyltransferase 5 (alpha (1,3) fucosyltransferase) |
| chr4_+_31998999 | 0.05 |
ENST00000513211.1
|
RP11-734I18.1
|
RP11-734I18.1 |
| chr9_-_98189055 | 0.05 |
ENST00000433644.2
|
RP11-435O5.2
|
RP11-435O5.2 |
| chr1_+_44514040 | 0.05 |
ENST00000431800.1
ENST00000437643.1 |
RP5-1198O20.4
|
RP5-1198O20.4 |
| chr10_-_14050522 | 0.05 |
ENST00000342409.2
|
FRMD4A
|
FERM domain containing 4A |
| chr13_+_108921977 | 0.05 |
ENST00000430559.1
ENST00000375887.4 |
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr18_+_61575200 | 0.05 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr8_-_124749609 | 0.05 |
ENST00000262219.6
ENST00000419625.1 |
ANXA13
|
annexin A13 |
| chr6_+_26156551 | 0.04 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr4_+_41258786 | 0.04 |
ENST00000503431.1
ENST00000284440.4 ENST00000508768.1 ENST00000512788.1 |
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr2_-_136288113 | 0.04 |
ENST00000401392.1
|
ZRANB3
|
zinc finger, RAN-binding domain containing 3 |
| chr10_+_102222798 | 0.04 |
ENST00000343737.5
|
WNT8B
|
wingless-type MMTV integration site family, member 8B |
| chr6_+_13272904 | 0.04 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr16_+_57653596 | 0.04 |
ENST00000568791.1
ENST00000570044.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr5_+_67586465 | 0.04 |
ENST00000336483.5
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr12_-_31881944 | 0.04 |
ENST00000537562.1
ENST00000537960.1 ENST00000536761.1 ENST00000542781.1 ENST00000457428.2 |
AMN1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae) |
| chr3_-_138048682 | 0.04 |
ENST00000383180.2
|
NME9
|
NME/NM23 family member 9 |
| chr3_+_41241596 | 0.04 |
ENST00000450969.1
|
CTNNB1
|
catenin (cadherin-associated protein), beta 1, 88kDa |
| chr16_+_20911174 | 0.04 |
ENST00000568663.1
|
LYRM1
|
LYR motif containing 1 |
| chr10_-_52008313 | 0.04 |
ENST00000329428.6
ENST00000395526.4 ENST00000447815.1 |
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 |
| chr7_+_77469439 | 0.04 |
ENST00000450574.1
ENST00000416283.2 ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr3_-_165555200 | 0.04 |
ENST00000479451.1
ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE
|
butyrylcholinesterase |
| chr7_-_5553369 | 0.04 |
ENST00000453700.3
ENST00000382368.3 |
FBXL18
|
F-box and leucine-rich repeat protein 18 |
| chr12_+_52056548 | 0.04 |
ENST00000545061.1
ENST00000355133.3 |
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit |
| chr3_-_69591727 | 0.04 |
ENST00000459638.1
|
FRMD4B
|
FERM domain containing 4B |
| chr3_-_185270383 | 0.04 |
ENST00000296252.4
|
LIPH
|
lipase, member H |
| chr9_+_108463234 | 0.04 |
ENST00000374688.1
|
TMEM38B
|
transmembrane protein 38B |
| chr6_+_148593425 | 0.04 |
ENST00000367469.1
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr11_+_86748863 | 0.04 |
ENST00000340353.7
|
TMEM135
|
transmembrane protein 135 |
| chr2_+_64069016 | 0.04 |
ENST00000488245.2
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr2_+_64073187 | 0.04 |
ENST00000491621.1
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr19_+_55897297 | 0.04 |
ENST00000431533.2
ENST00000428193.2 ENST00000558815.1 ENST00000560583.1 ENST00000560055.1 ENST00000559463.1 |
RPL28
|
ribosomal protein L28 |
| chr16_-_21863541 | 0.04 |
ENST00000543654.1
|
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr10_+_94352956 | 0.04 |
ENST00000260731.3
|
KIF11
|
kinesin family member 11 |
| chr12_-_25055177 | 0.04 |
ENST00000538118.1
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr17_+_61151306 | 0.04 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr1_+_948803 | 0.04 |
ENST00000379389.4
|
ISG15
|
ISG15 ubiquitin-like modifier |
| chr5_+_68860949 | 0.04 |
ENST00000507595.1
|
GTF2H2C
|
general transcription factor IIH, polypeptide 2C |
| chr18_-_52989525 | 0.04 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr8_+_38662960 | 0.04 |
ENST00000524193.1
|
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr11_+_126173647 | 0.04 |
ENST00000263579.4
|
DCPS
|
decapping enzyme, scavenger |
| chr14_+_93357749 | 0.04 |
ENST00000557689.1
|
RP11-862G15.1
|
RP11-862G15.1 |
| chr12_+_130646999 | 0.04 |
ENST00000539839.1
ENST00000229030.4 |
FZD10
|
frizzled family receptor 10 |
| chr12_+_72061563 | 0.04 |
ENST00000551238.1
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr19_-_19626351 | 0.04 |
ENST00000585580.3
|
TSSK6
|
testis-specific serine kinase 6 |
| chr11_+_27062502 | 0.04 |
ENST00000263182.3
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr19_-_22018966 | 0.04 |
ENST00000599906.1
ENST00000354959.4 |
ZNF43
|
zinc finger protein 43 |
| chr12_-_112443830 | 0.04 |
ENST00000550037.1
ENST00000549425.1 |
TMEM116
|
transmembrane protein 116 |
| chr15_-_83952071 | 0.04 |
ENST00000569704.1
|
BNC1
|
basonuclin 1 |
| chr2_+_172309634 | 0.04 |
ENST00000339506.3
|
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr3_-_47023455 | 0.04 |
ENST00000446836.1
ENST00000425441.1 |
CCDC12
|
coiled-coil domain containing 12 |
| chr2_+_242312264 | 0.04 |
ENST00000445489.1
|
FARP2
|
FERM, RhoGEF and pleckstrin domain protein 2 |
| chr8_+_118147498 | 0.04 |
ENST00000519688.1
ENST00000456015.2 |
SLC30A8
|
solute carrier family 30 (zinc transporter), member 8 |
| chr7_-_112430647 | 0.04 |
ENST00000312814.6
|
TMEM168
|
transmembrane protein 168 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD11_HOXA11
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.2 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.0 | 0.2 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.1 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.0 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.0 | GO:0009820 | alkaloid metabolic process(GO:0009820) |
| 0.0 | 0.0 | GO:0070839 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) divalent metal ion export(GO:0070839) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.0 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.1 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) UDP-glucuronate metabolic process(GO:0046398) |
| 0.0 | 0.0 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.0 | GO:1902997 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.0 | 0.0 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.0 | GO:1904499 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.0 | 0.0 | GO:0042214 | terpene metabolic process(GO:0042214) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.0 | GO:0031592 | centrosomal corona(GO:0031592) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.1 | 0.2 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.0 | 0.2 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0005427 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.2 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.0 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.0 | 0.1 | GO:0035529 | phosphodiesterase I activity(GO:0004528) NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.0 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.0 | GO:0004423 | iduronate-2-sulfatase activity(GO:0004423) |
| 0.0 | 0.0 | GO:0033265 | acetylcholinesterase activity(GO:0003990) choline binding(GO:0033265) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.0 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |