Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
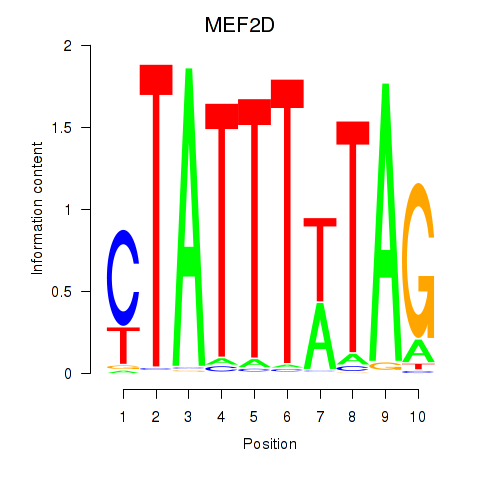
Results for MEF2D_MEF2A
Z-value: 0.40
Transcription factors associated with MEF2D_MEF2A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MEF2D
|
ENSG00000116604.13 | myocyte enhancer factor 2D |
|
MEF2A
|
ENSG00000068305.13 | myocyte enhancer factor 2A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MEF2A | hg19_v2_chr15_+_100106244_100106292 | -0.72 | 1.1e-01 | Click! |
| MEF2D | hg19_v2_chr1_-_156460391_156460417 | 0.56 | 2.5e-01 | Click! |
Activity profile of MEF2D_MEF2A motif
Sorted Z-values of MEF2D_MEF2A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_37821593 | 0.45 |
ENST00000578283.1
|
TCAP
|
titin-cap |
| chr22_-_30642728 | 0.37 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr8_+_40010989 | 0.32 |
ENST00000315792.3
|
C8orf4
|
chromosome 8 open reading frame 4 |
| chr14_+_103589789 | 0.28 |
ENST00000558056.1
ENST00000560869.1 |
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2 |
| chr22_+_25595817 | 0.26 |
ENST00000215855.2
ENST00000404334.1 |
CRYBB3
|
crystallin, beta B3 |
| chr6_-_27806117 | 0.25 |
ENST00000330180.2
|
HIST1H2AK
|
histone cluster 1, H2ak |
| chr7_-_41742697 | 0.25 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr8_-_73793975 | 0.21 |
ENST00000523881.1
|
RP11-1145L24.1
|
RP11-1145L24.1 |
| chr6_+_26020672 | 0.19 |
ENST00000357647.3
|
HIST1H3A
|
histone cluster 1, H3a |
| chr7_+_18536090 | 0.19 |
ENST00000441986.1
|
HDAC9
|
histone deacetylase 9 |
| chr14_-_103989033 | 0.18 |
ENST00000553878.1
ENST00000557530.1 |
CKB
|
creatine kinase, brain |
| chr1_-_204436344 | 0.17 |
ENST00000367184.2
|
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr17_+_80186908 | 0.16 |
ENST00000582743.1
ENST00000578684.1 ENST00000577650.1 ENST00000582715.1 |
SLC16A3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr17_+_7758374 | 0.16 |
ENST00000301599.6
ENST00000574668.1 |
TMEM88
|
transmembrane protein 88 |
| chr1_+_1846519 | 0.16 |
ENST00000378604.3
|
CALML6
|
calmodulin-like 6 |
| chr17_-_79900255 | 0.15 |
ENST00000330655.3
ENST00000582198.1 |
MYADML2
PYCR1
|
myeloid-associated differentiation marker-like 2 pyrroline-5-carboxylate reductase 1 |
| chr17_+_6918064 | 0.15 |
ENST00000546760.1
ENST00000552402.1 |
C17orf49
|
chromosome 17 open reading frame 49 |
| chr11_+_65082289 | 0.15 |
ENST00000279249.2
|
CDC42EP2
|
CDC42 effector protein (Rho GTPase binding) 2 |
| chr19_-_18209626 | 0.15 |
ENST00000598019.2
ENST00000594176.1 ENST00000600835.2 |
IL12RB1
|
interleukin 12 receptor, beta 1 |
| chr3_+_99357319 | 0.15 |
ENST00000452013.1
ENST00000261037.3 ENST00000273342.4 |
COL8A1
|
collagen, type VIII, alpha 1 |
| chr12_-_57644952 | 0.14 |
ENST00000554578.1
ENST00000546246.2 ENST00000553489.1 ENST00000332782.2 |
STAC3
|
SH3 and cysteine rich domain 3 |
| chr17_+_6918093 | 0.14 |
ENST00000439424.2
|
C17orf49
|
chromosome 17 open reading frame 49 |
| chr19_+_16607122 | 0.14 |
ENST00000221671.3
ENST00000594035.1 ENST00000599550.1 ENST00000594813.1 |
C19orf44
|
chromosome 19 open reading frame 44 |
| chr17_-_6917755 | 0.14 |
ENST00000593646.1
|
AC040977.1
|
Uncharacterized protein |
| chr15_+_74509530 | 0.14 |
ENST00000321288.5
|
CCDC33
|
coiled-coil domain containing 33 |
| chr14_-_21493123 | 0.13 |
ENST00000556147.1
ENST00000554489.1 ENST00000555657.1 ENST00000557274.1 ENST00000555158.1 ENST00000554833.1 ENST00000555384.1 ENST00000556420.1 ENST00000554893.1 ENST00000553503.1 ENST00000555733.1 ENST00000553867.1 ENST00000397856.3 ENST00000397855.3 ENST00000556008.1 ENST00000557182.1 ENST00000554483.1 ENST00000556688.1 ENST00000397853.3 ENST00000556329.2 ENST00000554143.1 ENST00000397851.2 ENST00000555142.1 ENST00000557676.1 ENST00000556924.1 |
NDRG2
|
NDRG family member 2 |
| chr16_+_12059091 | 0.13 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr15_+_91446961 | 0.13 |
ENST00000559965.1
|
MAN2A2
|
mannosidase, alpha, class 2A, member 2 |
| chr3_-_57326704 | 0.12 |
ENST00000487349.1
ENST00000389601.3 |
ASB14
|
ankyrin repeat and SOCS box containing 14 |
| chr8_-_23282820 | 0.12 |
ENST00000520871.1
|
LOXL2
|
lysyl oxidase-like 2 |
| chr16_+_30075783 | 0.12 |
ENST00000412304.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr14_-_21493649 | 0.12 |
ENST00000553442.1
ENST00000555869.1 ENST00000556457.1 ENST00000397844.2 ENST00000554415.1 |
NDRG2
|
NDRG family member 2 |
| chr11_-_7904464 | 0.12 |
ENST00000502624.2
ENST00000527565.1 |
RP11-35J10.5
|
RP11-35J10.5 |
| chr9_-_33402506 | 0.12 |
ENST00000377425.4
ENST00000537089.1 ENST00000297988.1 ENST00000539936.1 ENST00000541274.1 |
AQP7
|
aquaporin 7 |
| chr16_+_30075595 | 0.11 |
ENST00000563060.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr16_+_30076052 | 0.11 |
ENST00000563987.1
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr7_+_18535321 | 0.11 |
ENST00000413380.1
ENST00000430454.1 |
HDAC9
|
histone deacetylase 9 |
| chr7_+_18126557 | 0.11 |
ENST00000417496.2
|
HDAC9
|
histone deacetylase 9 |
| chr8_-_23282797 | 0.10 |
ENST00000524144.1
|
LOXL2
|
lysyl oxidase-like 2 |
| chr14_-_21493884 | 0.10 |
ENST00000556974.1
ENST00000554419.1 ENST00000298687.5 ENST00000397858.1 ENST00000360463.3 ENST00000350792.3 ENST00000397847.2 |
NDRG2
|
NDRG family member 2 |
| chr3_-_156878482 | 0.10 |
ENST00000295925.4
|
CCNL1
|
cyclin L1 |
| chr12_+_116985896 | 0.10 |
ENST00000547114.1
|
RP11-809C9.2
|
RP11-809C9.2 |
| chr5_-_150521192 | 0.10 |
ENST00000523714.1
ENST00000521749.1 |
ANXA6
|
annexin A6 |
| chr7_-_134143841 | 0.10 |
ENST00000285930.4
|
AKR1B1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr12_-_22094336 | 0.10 |
ENST00000326684.4
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr5_+_132009675 | 0.10 |
ENST00000231449.2
ENST00000350025.2 |
IL4
|
interleukin 4 |
| chr22_-_51017084 | 0.10 |
ENST00000360719.2
ENST00000457250.1 ENST00000440709.1 |
CPT1B
|
carnitine palmitoyltransferase 1B (muscle) |
| chr11_-_104480019 | 0.09 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr3_-_48598547 | 0.09 |
ENST00000536104.1
ENST00000452531.1 |
PFKFB4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr9_-_136344237 | 0.09 |
ENST00000432868.1
ENST00000371899.4 |
SLC2A6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr17_-_48277552 | 0.09 |
ENST00000507689.1
|
COL1A1
|
collagen, type I, alpha 1 |
| chr10_+_104178946 | 0.09 |
ENST00000432590.1
|
FBXL15
|
F-box and leucine-rich repeat protein 15 |
| chr7_-_44105158 | 0.09 |
ENST00000297283.3
|
PGAM2
|
phosphoglycerate mutase 2 (muscle) |
| chr12_-_49581152 | 0.09 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr10_-_3827371 | 0.09 |
ENST00000469435.1
|
KLF6
|
Kruppel-like factor 6 |
| chr22_-_51016846 | 0.09 |
ENST00000312108.7
ENST00000395650.2 |
CPT1B
|
carnitine palmitoyltransferase 1B (muscle) |
| chr6_+_131571535 | 0.08 |
ENST00000474850.2
|
AKAP7
|
A kinase (PRKA) anchor protein 7 |
| chr10_-_74114714 | 0.08 |
ENST00000338820.3
ENST00000394903.2 ENST00000444643.2 |
DNAJB12
|
DnaJ (Hsp40) homolog, subfamily B, member 12 |
| chr7_+_150758642 | 0.08 |
ENST00000488420.1
|
SLC4A2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr1_+_114522049 | 0.08 |
ENST00000369551.1
ENST00000320334.4 |
OLFML3
|
olfactomedin-like 3 |
| chr15_+_59730348 | 0.08 |
ENST00000288228.5
ENST00000559628.1 ENST00000557914.1 ENST00000560474.1 |
FAM81A
|
family with sequence similarity 81, member A |
| chr10_+_77056181 | 0.08 |
ENST00000526759.1
ENST00000533822.1 |
ZNF503-AS1
|
ZNF503 antisense RNA 1 |
| chr16_+_30075463 | 0.08 |
ENST00000562168.1
ENST00000569545.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr1_+_26348259 | 0.08 |
ENST00000374280.3
|
EXTL1
|
exostosin-like glycosyltransferase 1 |
| chr5_+_66300446 | 0.08 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr7_+_143013198 | 0.08 |
ENST00000343257.2
|
CLCN1
|
chloride channel, voltage-sensitive 1 |
| chr6_+_15401075 | 0.08 |
ENST00000541660.1
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr6_-_6007200 | 0.08 |
ENST00000244766.2
|
NRN1
|
neuritin 1 |
| chr10_-_75415825 | 0.08 |
ENST00000394810.2
|
SYNPO2L
|
synaptopodin 2-like |
| chr6_+_26104104 | 0.07 |
ENST00000377803.2
|
HIST1H4C
|
histone cluster 1, H4c |
| chr3_+_108541545 | 0.07 |
ENST00000295756.6
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr17_-_1389228 | 0.07 |
ENST00000438665.2
|
MYO1C
|
myosin IC |
| chr3_-_148939598 | 0.07 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr5_-_141030943 | 0.07 |
ENST00000522783.1
ENST00000519800.1 ENST00000435817.2 |
FCHSD1
|
FCH and double SH3 domains 1 |
| chr2_-_190044480 | 0.07 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr7_+_18535346 | 0.07 |
ENST00000405010.3
ENST00000406451.4 ENST00000428307.2 |
HDAC9
|
histone deacetylase 9 |
| chr20_+_61147651 | 0.07 |
ENST00000370527.3
ENST00000370524.2 |
C20orf166
|
chromosome 20 open reading frame 166 |
| chr21_+_17909594 | 0.07 |
ENST00000441820.1
ENST00000602280.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr9_+_100263912 | 0.07 |
ENST00000259365.4
|
TMOD1
|
tropomodulin 1 |
| chr1_-_63782888 | 0.07 |
ENST00000436475.2
|
LINC00466
|
long intergenic non-protein coding RNA 466 |
| chr17_-_42200996 | 0.07 |
ENST00000587135.1
ENST00000225983.6 ENST00000393622.2 ENST00000588703.1 |
HDAC5
|
histone deacetylase 5 |
| chr20_+_48807351 | 0.07 |
ENST00000303004.3
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta |
| chr1_+_156095951 | 0.07 |
ENST00000448611.2
ENST00000368297.1 |
LMNA
|
lamin A/C |
| chr9_-_136344197 | 0.06 |
ENST00000414172.1
ENST00000371897.4 |
SLC2A6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr12_-_111358372 | 0.06 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr2_-_31637560 | 0.06 |
ENST00000379416.3
|
XDH
|
xanthine dehydrogenase |
| chr1_+_183605222 | 0.06 |
ENST00000536277.1
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1 |
| chrX_-_63450480 | 0.06 |
ENST00000362002.2
|
ASB12
|
ankyrin repeat and SOCS box containing 12 |
| chr9_+_116327326 | 0.06 |
ENST00000342620.5
|
RGS3
|
regulator of G-protein signaling 3 |
| chr2_-_27938593 | 0.06 |
ENST00000379677.2
|
AC074091.13
|
Uncharacterized protein |
| chr7_+_120629653 | 0.06 |
ENST00000450913.2
ENST00000340646.5 |
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr6_+_106535455 | 0.06 |
ENST00000424894.1
|
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr6_+_136172820 | 0.06 |
ENST00000308191.6
|
PDE7B
|
phosphodiesterase 7B |
| chr7_+_18535893 | 0.06 |
ENST00000432645.2
ENST00000441542.2 |
HDAC9
|
histone deacetylase 9 |
| chr5_+_66300464 | 0.06 |
ENST00000436277.1
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr15_+_78558523 | 0.06 |
ENST00000446172.2
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4 |
| chr4_+_160025300 | 0.06 |
ENST00000505478.1
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr10_-_75410771 | 0.06 |
ENST00000372873.4
|
SYNPO2L
|
synaptopodin 2-like |
| chr8_-_71157595 | 0.06 |
ENST00000519724.1
|
NCOA2
|
nuclear receptor coactivator 2 |
| chr7_-_22259845 | 0.06 |
ENST00000420196.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr3_+_193853927 | 0.06 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr11_+_128563652 | 0.06 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr5_-_115910630 | 0.06 |
ENST00000343348.6
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr7_-_55583740 | 0.06 |
ENST00000453256.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr2_-_179914760 | 0.06 |
ENST00000420890.2
ENST00000409284.1 ENST00000443758.1 ENST00000446116.1 |
CCDC141
|
coiled-coil domain containing 141 |
| chr17_-_1389419 | 0.06 |
ENST00000575158.1
|
MYO1C
|
myosin IC |
| chr1_-_116311402 | 0.06 |
ENST00000261448.5
|
CASQ2
|
calsequestrin 2 (cardiac muscle) |
| chr9_+_215158 | 0.06 |
ENST00000479404.1
|
DOCK8
|
dedicator of cytokinesis 8 |
| chrX_+_18725758 | 0.06 |
ENST00000472826.1
ENST00000544635.1 ENST00000496075.2 |
PPEF1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr9_+_130565147 | 0.06 |
ENST00000373247.2
ENST00000373245.1 ENST00000393706.2 ENST00000373228.1 |
FPGS
|
folylpolyglutamate synthase |
| chr17_-_4871085 | 0.05 |
ENST00000575142.1
ENST00000206020.3 |
SPAG7
|
sperm associated antigen 7 |
| chr10_+_71561630 | 0.05 |
ENST00000398974.3
ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr1_-_27240455 | 0.05 |
ENST00000254227.3
|
NR0B2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr15_-_93353028 | 0.05 |
ENST00000557398.2
|
FAM174B
|
family with sequence similarity 174, member B |
| chr6_+_53948328 | 0.05 |
ENST00000370876.2
|
MLIP
|
muscular LMNA-interacting protein |
| chr12_-_16760195 | 0.05 |
ENST00000546281.1
ENST00000537757.1 |
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr17_-_41132088 | 0.05 |
ENST00000591916.1
ENST00000451885.2 ENST00000454303.1 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr1_+_36621697 | 0.05 |
ENST00000373150.4
ENST00000373151.2 |
MAP7D1
|
MAP7 domain containing 1 |
| chr5_-_141704566 | 0.05 |
ENST00000344120.4
ENST00000434127.2 |
SPRY4
|
sprouty homolog 4 (Drosophila) |
| chr20_-_44519839 | 0.05 |
ENST00000372518.4
|
NEURL2
|
neuralized E3 ubiquitin protein ligase 2 |
| chrM_+_7586 | 0.05 |
ENST00000361739.1
|
MT-CO2
|
mitochondrially encoded cytochrome c oxidase II |
| chr12_-_16760021 | 0.05 |
ENST00000540445.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr10_-_49860525 | 0.05 |
ENST00000435790.2
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr16_+_50313426 | 0.05 |
ENST00000569265.1
|
ADCY7
|
adenylate cyclase 7 |
| chr6_-_41703952 | 0.05 |
ENST00000358871.2
ENST00000403298.4 |
TFEB
|
transcription factor EB |
| chr19_+_35630344 | 0.05 |
ENST00000455515.2
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr1_-_59249732 | 0.05 |
ENST00000371222.2
|
JUN
|
jun proto-oncogene |
| chr1_+_171060018 | 0.05 |
ENST00000367755.4
ENST00000392085.2 ENST00000542847.1 ENST00000538429.1 ENST00000479749.1 |
FMO3
|
flavin containing monooxygenase 3 |
| chr3_+_113616317 | 0.05 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr14_-_102704783 | 0.05 |
ENST00000522534.1
|
MOK
|
MOK protein kinase |
| chr11_+_45918092 | 0.05 |
ENST00000395629.2
|
MAPK8IP1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chrX_-_138304939 | 0.05 |
ENST00000448673.1
|
FGF13
|
fibroblast growth factor 13 |
| chr17_-_74380565 | 0.05 |
ENST00000442767.1
ENST00000423915.1 |
PRPSAP1
|
phosphoribosyl pyrophosphate synthetase-associated protein 1 |
| chr10_+_71562180 | 0.05 |
ENST00000517713.1
ENST00000522165.1 ENST00000520133.1 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr22_-_31328881 | 0.05 |
ENST00000445980.1
|
MORC2
|
MORC family CW-type zinc finger 2 |
| chr13_+_76378305 | 0.04 |
ENST00000526371.1
ENST00000526528.1 |
LMO7
|
LIM domain 7 |
| chr12_-_22094159 | 0.04 |
ENST00000538350.1
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr19_+_35629702 | 0.04 |
ENST00000351325.4
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr15_+_73976715 | 0.04 |
ENST00000558689.1
ENST00000560786.2 ENST00000561213.1 ENST00000563584.1 ENST00000561416.1 |
CD276
|
CD276 molecule |
| chr8_-_42234745 | 0.04 |
ENST00000220812.2
|
DKK4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chr9_+_8858102 | 0.04 |
ENST00000447950.1
ENST00000430766.1 |
RP11-75C9.1
|
RP11-75C9.1 |
| chr19_-_40324767 | 0.04 |
ENST00000601972.1
ENST00000430012.2 ENST00000323039.5 ENST00000348817.3 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr9_+_97766469 | 0.04 |
ENST00000433691.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr6_-_169654139 | 0.04 |
ENST00000366787.3
|
THBS2
|
thrombospondin 2 |
| chr17_-_41132410 | 0.04 |
ENST00000409446.3
ENST00000453594.1 ENST00000409399.1 ENST00000421990.2 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr22_+_31489344 | 0.04 |
ENST00000404574.1
|
SMTN
|
smoothelin |
| chr9_-_215744 | 0.04 |
ENST00000382387.2
|
C9orf66
|
chromosome 9 open reading frame 66 |
| chr2_-_220117867 | 0.04 |
ENST00000456818.1
ENST00000447205.1 |
TUBA4A
|
tubulin, alpha 4a |
| chr3_+_188664988 | 0.04 |
ENST00000433971.1
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr6_-_123958051 | 0.04 |
ENST00000546248.1
|
TRDN
|
triadin |
| chr3_-_148939835 | 0.04 |
ENST00000264613.6
|
CP
|
ceruloplasmin (ferroxidase) |
| chr10_+_14880157 | 0.04 |
ENST00000378372.3
|
HSPA14
|
heat shock 70kDa protein 14 |
| chr20_+_42574317 | 0.04 |
ENST00000358131.5
|
TOX2
|
TOX high mobility group box family member 2 |
| chr4_-_76823681 | 0.04 |
ENST00000286719.7
|
PPEF2
|
protein phosphatase, EF-hand calcium binding domain 2 |
| chr6_+_7108210 | 0.04 |
ENST00000467782.1
ENST00000334984.6 ENST00000349384.6 |
RREB1
|
ras responsive element binding protein 1 |
| chr3_-_192445289 | 0.04 |
ENST00000430714.1
ENST00000418610.1 ENST00000448795.1 ENST00000445105.2 |
FGF12
|
fibroblast growth factor 12 |
| chr17_-_41132010 | 0.04 |
ENST00000409103.1
ENST00000360221.4 |
PTGES3L-AARSD1
|
PTGES3L-AARSD1 readthrough |
| chr2_+_66918558 | 0.04 |
ENST00000435389.1
ENST00000428590.1 ENST00000412944.1 |
AC007392.3
|
AC007392.3 |
| chr1_-_12677714 | 0.04 |
ENST00000376223.2
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr4_+_186064395 | 0.04 |
ENST00000281456.6
|
SLC25A4
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 |
| chr10_+_71561649 | 0.03 |
ENST00000398978.3
ENST00000354547.3 ENST00000357811.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr3_-_52486841 | 0.03 |
ENST00000496590.1
|
TNNC1
|
troponin C type 1 (slow) |
| chr1_+_170633047 | 0.03 |
ENST00000239461.6
ENST00000497230.2 |
PRRX1
|
paired related homeobox 1 |
| chr1_-_31845914 | 0.03 |
ENST00000373713.2
|
FABP3
|
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) |
| chr4_+_89514516 | 0.03 |
ENST00000452979.1
|
HERC3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr4_-_170192000 | 0.03 |
ENST00000502315.1
|
SH3RF1
|
SH3 domain containing ring finger 1 |
| chr6_-_2751146 | 0.03 |
ENST00000268446.5
ENST00000274643.7 |
MYLK4
|
myosin light chain kinase family, member 4 |
| chr19_-_40324255 | 0.03 |
ENST00000593685.1
ENST00000600611.1 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr10_+_102758105 | 0.03 |
ENST00000429732.1
|
LZTS2
|
leucine zipper, putative tumor suppressor 2 |
| chr2_+_233497931 | 0.03 |
ENST00000264059.3
|
EFHD1
|
EF-hand domain family, member D1 |
| chr9_+_70971815 | 0.03 |
ENST00000396392.1
ENST00000396396.1 |
PGM5
|
phosphoglucomutase 5 |
| chr17_-_36891830 | 0.03 |
ENST00000578487.1
|
PCGF2
|
polycomb group ring finger 2 |
| chr17_+_4855053 | 0.03 |
ENST00000518175.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr2_+_220143989 | 0.03 |
ENST00000336576.5
|
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr16_-_18923035 | 0.03 |
ENST00000563836.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr15_-_38852251 | 0.03 |
ENST00000558432.1
|
RASGRP1
|
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
| chr5_+_140552218 | 0.03 |
ENST00000231137.3
|
PCDHB7
|
protocadherin beta 7 |
| chr11_-_18034701 | 0.03 |
ENST00000265965.5
|
SERGEF
|
secretion regulating guanine nucleotide exchange factor |
| chr3_+_8543393 | 0.03 |
ENST00000157600.3
ENST00000415597.1 ENST00000535732.1 |
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr4_-_23891658 | 0.03 |
ENST00000507380.1
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chr21_-_34185944 | 0.03 |
ENST00000479548.1
|
C21orf62
|
chromosome 21 open reading frame 62 |
| chr6_-_26124138 | 0.03 |
ENST00000314332.5
ENST00000396984.1 |
HIST1H2BC
|
histone cluster 1, H2bc |
| chr5_+_66124590 | 0.03 |
ENST00000490016.2
ENST00000403666.1 ENST00000450827.1 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr20_+_32581525 | 0.03 |
ENST00000246194.3
ENST00000413297.1 |
RALY
|
RALY heterogeneous nuclear ribonucleoprotein |
| chr1_+_28586006 | 0.03 |
ENST00000253063.3
|
SESN2
|
sestrin 2 |
| chr10_-_104178857 | 0.03 |
ENST00000020673.5
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr10_+_133753533 | 0.03 |
ENST00000422256.2
|
PPP2R2D
|
protein phosphatase 2, regulatory subunit B, delta |
| chr1_+_228395755 | 0.03 |
ENST00000284548.11
ENST00000570156.2 ENST00000422127.1 ENST00000366707.4 ENST00000366709.4 |
OBSCN
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr1_-_116383322 | 0.03 |
ENST00000429731.1
|
NHLH2
|
nescient helix loop helix 2 |
| chr2_+_33683109 | 0.03 |
ENST00000437184.1
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr17_-_27949911 | 0.03 |
ENST00000492276.2
ENST00000345068.5 ENST00000584602.1 |
CORO6
|
coronin 6 |
| chr9_-_119162885 | 0.03 |
ENST00000445861.2
|
PAPPA-AS1
|
PAPPA antisense RNA 1 |
| chr1_+_150254936 | 0.03 |
ENST00000447007.1
ENST00000369095.1 ENST00000369094.1 |
C1orf51
|
chromosome 1 open reading frame 51 |
| chr12_-_45269430 | 0.03 |
ENST00000395487.2
|
NELL2
|
NEL-like 2 (chicken) |
| chr13_-_67802549 | 0.03 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr6_-_135424186 | 0.03 |
ENST00000529882.1
|
HBS1L
|
HBS1-like (S. cerevisiae) |
| chr3_+_36421971 | 0.03 |
ENST00000457375.2
ENST00000434649.1 |
STAC
|
SH3 and cysteine rich domain |
| chr6_-_76203454 | 0.03 |
ENST00000237172.7
|
FILIP1
|
filamin A interacting protein 1 |
| chr1_+_81106951 | 0.03 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr2_+_33661382 | 0.03 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
Network of associatons between targets according to the STRING database.
First level regulatory network of MEF2D_MEF2A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 0.3 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.4 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.2 | GO:0018277 | protein deamination(GO:0018277) |
| 0.0 | 0.2 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.3 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.5 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.2 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.2 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.4 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.0 | 0.1 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:2000974 | negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.0 | 0.1 | GO:0061485 | memory T cell proliferation(GO:0061485) |
| 0.0 | 0.1 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.0 | 0.0 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.1 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.1 | GO:0042022 | interleukin-12 receptor complex(GO:0042022) |
| 0.0 | 0.1 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 0.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.1 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 0.4 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 0.3 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0016517 | interleukin-12 receptor activity(GO:0016517) |
| 0.0 | 0.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.2 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.0 | 0.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.1 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0008841 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) dihydrofolate synthase activity(GO:0008841) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.6 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.2 | GO:0035005 | lipid kinase activity(GO:0001727) 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.0 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |