Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
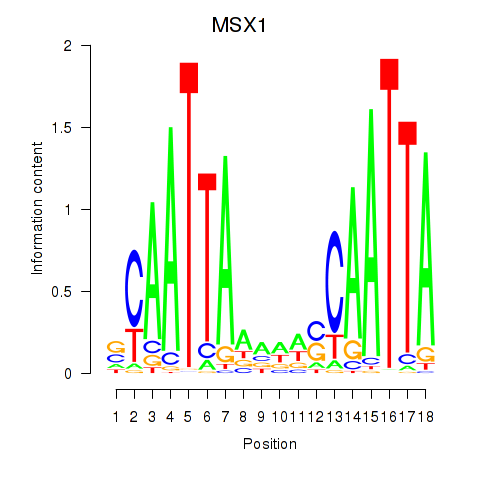
Results for MSX1
Z-value: 0.86
Transcription factors associated with MSX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MSX1
|
ENSG00000163132.6 | msh homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MSX1 | hg19_v2_chr4_+_4861385_4861398 | 0.12 | 8.2e-01 | Click! |
Activity profile of MSX1 motif
Sorted Z-values of MSX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_119669160 | 1.01 |
ENST00000514240.1
|
CTC-552D5.1
|
CTC-552D5.1 |
| chr7_+_106415457 | 0.79 |
ENST00000490162.2
ENST00000470135.1 |
RP5-884M6.1
|
RP5-884M6.1 |
| chr8_-_25281747 | 0.75 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr21_+_17792672 | 0.63 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr19_-_45457264 | 0.60 |
ENST00000591646.1
|
CTB-129P6.11
|
Uncharacterized protein |
| chr17_+_74729060 | 0.57 |
ENST00000587459.1
|
RP11-318A15.7
|
Uncharacterized protein |
| chr5_-_138739739 | 0.51 |
ENST00000514983.1
ENST00000507779.2 ENST00000451821.2 ENST00000450845.2 ENST00000509959.1 ENST00000302091.5 |
SPATA24
|
spermatogenesis associated 24 |
| chr4_+_9446156 | 0.41 |
ENST00000334879.1
|
DEFB131
|
defensin, beta 131 |
| chr22_-_32767017 | 0.33 |
ENST00000400234.1
|
RFPL3S
|
RFPL3 antisense |
| chr10_-_61899124 | 0.31 |
ENST00000373815.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr2_+_196313239 | 0.31 |
ENST00000413290.1
|
AC064834.1
|
AC064834.1 |
| chr6_+_28249299 | 0.30 |
ENST00000405948.2
|
PGBD1
|
piggyBac transposable element derived 1 |
| chr6_+_26020672 | 0.29 |
ENST00000357647.3
|
HIST1H3A
|
histone cluster 1, H3a |
| chr21_+_39668478 | 0.29 |
ENST00000398927.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr16_-_71518504 | 0.28 |
ENST00000565100.2
|
ZNF19
|
zinc finger protein 19 |
| chr15_+_77713222 | 0.27 |
ENST00000558176.1
|
HMG20A
|
high mobility group 20A |
| chr13_-_81801115 | 0.27 |
ENST00000567258.1
|
LINC00564
|
long intergenic non-protein coding RNA 564 |
| chr7_-_16840820 | 0.27 |
ENST00000450569.1
|
AGR2
|
anterior gradient 2 |
| chr11_-_85376121 | 0.26 |
ENST00000527447.1
|
CREBZF
|
CREB/ATF bZIP transcription factor |
| chr19_+_3762645 | 0.26 |
ENST00000330133.4
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr21_+_33671264 | 0.26 |
ENST00000339944.4
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr19_+_3762703 | 0.25 |
ENST00000589174.1
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr7_-_105332084 | 0.25 |
ENST00000472195.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr14_+_101293687 | 0.23 |
ENST00000455286.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr17_-_47786375 | 0.23 |
ENST00000511657.1
|
SLC35B1
|
solute carrier family 35, member B1 |
| chr4_+_71384300 | 0.22 |
ENST00000504451.1
|
AMTN
|
amelotin |
| chr4_-_120243545 | 0.22 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr20_+_5731027 | 0.21 |
ENST00000378979.4
ENST00000303142.6 |
C20orf196
|
chromosome 20 open reading frame 196 |
| chrX_+_1710484 | 0.21 |
ENST00000313871.3
ENST00000381261.3 |
AKAP17A
|
A kinase (PRKA) anchor protein 17A |
| chr7_+_16793160 | 0.21 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr14_-_73997901 | 0.20 |
ENST00000557603.1
ENST00000556455.1 |
HEATR4
|
HEAT repeat containing 4 |
| chr12_+_9066472 | 0.20 |
ENST00000538657.1
|
PHC1
|
polyhomeotic homolog 1 (Drosophila) |
| chr3_-_21792838 | 0.20 |
ENST00000281523.2
|
ZNF385D
|
zinc finger protein 385D |
| chr3_-_180397256 | 0.20 |
ENST00000442201.2
|
CCDC39
|
coiled-coil domain containing 39 |
| chr8_-_121824374 | 0.20 |
ENST00000517992.1
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
| chr9_-_70465758 | 0.20 |
ENST00000489273.1
|
CBWD5
|
COBW domain containing 5 |
| chr12_-_10978957 | 0.20 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr5_+_121647386 | 0.19 |
ENST00000542191.1
ENST00000506272.1 ENST00000508681.1 ENST00000509154.2 |
SNCAIP
|
synuclein, alpha interacting protein |
| chr6_-_109776901 | 0.18 |
ENST00000431946.1
|
MICAL1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr14_-_101295407 | 0.18 |
ENST00000596284.1
|
AL117190.2
|
AL117190.2 |
| chr2_-_219031709 | 0.17 |
ENST00000295683.2
|
CXCR1
|
chemokine (C-X-C motif) receptor 1 |
| chr6_+_118869452 | 0.16 |
ENST00000357525.5
|
PLN
|
phospholamban |
| chr7_+_141408153 | 0.16 |
ENST00000397541.2
|
WEE2
|
WEE1 homolog 2 (S. pombe) |
| chr22_+_51176624 | 0.16 |
ENST00000216139.5
ENST00000529621.1 |
ACR
|
acrosin |
| chr6_+_108616243 | 0.15 |
ENST00000421954.1
|
LACE1
|
lactation elevated 1 |
| chr9_+_44868935 | 0.15 |
ENST00000448436.2
|
RP11-160N1.10
|
RP11-160N1.10 |
| chr19_+_18303992 | 0.15 |
ENST00000599612.2
|
MPV17L2
|
MPV17 mitochondrial membrane protein-like 2 |
| chr16_-_66583994 | 0.15 |
ENST00000564917.1
|
TK2
|
thymidine kinase 2, mitochondrial |
| chr11_-_102401469 | 0.15 |
ENST00000260227.4
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr3_+_187957646 | 0.15 |
ENST00000457242.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr13_-_52733980 | 0.15 |
ENST00000339406.3
|
NEK3
|
NIMA-related kinase 3 |
| chr3_-_47555167 | 0.15 |
ENST00000296149.4
|
ELP6
|
elongator acetyltransferase complex subunit 6 |
| chr14_+_50291993 | 0.15 |
ENST00000595378.1
|
AL627171.2
|
HCG1786899; PRO2610; Uncharacterized protein |
| chr12_+_105380073 | 0.14 |
ENST00000552951.1
ENST00000280749.5 |
C12orf45
|
chromosome 12 open reading frame 45 |
| chr18_-_70532906 | 0.14 |
ENST00000299430.2
ENST00000397929.1 |
NETO1
|
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr4_-_186696425 | 0.14 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr2_+_242498135 | 0.14 |
ENST00000318407.3
|
BOK
|
BCL2-related ovarian killer |
| chr6_+_130339710 | 0.14 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr19_-_39805976 | 0.14 |
ENST00000248668.4
|
LRFN1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr16_+_28985542 | 0.14 |
ENST00000567771.1
ENST00000568388.1 |
SPNS1
|
spinster homolog 1 (Drosophila) |
| chr17_-_55162360 | 0.13 |
ENST00000576871.1
ENST00000576313.1 |
RP11-166P13.3
|
RP11-166P13.3 |
| chr5_-_177207634 | 0.13 |
ENST00000513554.1
ENST00000440605.3 |
FAM153A
|
family with sequence similarity 153, member A |
| chr11_-_128894053 | 0.13 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr17_+_72931876 | 0.13 |
ENST00000328801.4
|
OTOP3
|
otopetrin 3 |
| chr12_-_52604607 | 0.13 |
ENST00000551894.1
ENST00000553017.1 |
C12orf80
|
chromosome 12 open reading frame 80 |
| chr5_+_121465207 | 0.13 |
ENST00000296600.4
|
ZNF474
|
zinc finger protein 474 |
| chr19_+_107471 | 0.12 |
ENST00000585993.1
|
OR4F17
|
olfactory receptor, family 4, subfamily F, member 17 |
| chr5_+_140220769 | 0.12 |
ENST00000531613.1
ENST00000378123.3 |
PCDHA8
|
protocadherin alpha 8 |
| chrX_+_23928500 | 0.12 |
ENST00000435707.1
|
CXorf58
|
chromosome X open reading frame 58 |
| chr13_-_52733670 | 0.12 |
ENST00000550841.1
|
NEK3
|
NIMA-related kinase 3 |
| chr6_+_28092338 | 0.12 |
ENST00000340487.4
|
ZSCAN16
|
zinc finger and SCAN domain containing 16 |
| chr6_-_32908765 | 0.12 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr6_-_150212029 | 0.12 |
ENST00000529948.1
ENST00000357183.4 ENST00000367363.3 |
RAET1E
|
retinoic acid early transcript 1E |
| chr9_+_12695702 | 0.12 |
ENST00000381136.2
|
TYRP1
|
tyrosinase-related protein 1 |
| chr11_+_58695096 | 0.12 |
ENST00000525608.1
ENST00000526351.1 |
GLYATL1
|
glycine-N-acyltransferase-like 1 |
| chr4_-_186570679 | 0.12 |
ENST00000451974.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr6_+_28249332 | 0.11 |
ENST00000259883.3
|
PGBD1
|
piggyBac transposable element derived 1 |
| chr1_-_54405773 | 0.11 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr2_-_31361543 | 0.11 |
ENST00000349752.5
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr6_-_109777128 | 0.11 |
ENST00000358807.3
ENST00000358577.3 |
MICAL1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr21_+_30502806 | 0.11 |
ENST00000399928.1
ENST00000399926.1 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr16_-_66584059 | 0.11 |
ENST00000417693.3
ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr17_+_55162453 | 0.11 |
ENST00000575322.1
ENST00000337714.3 ENST00000314126.3 |
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr5_-_147162078 | 0.11 |
ENST00000507386.1
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2 |
| chr21_+_33671160 | 0.10 |
ENST00000303645.5
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr4_+_119809984 | 0.10 |
ENST00000307142.4
ENST00000448416.2 ENST00000429713.2 |
SYNPO2
|
synaptopodin 2 |
| chr14_+_68086515 | 0.10 |
ENST00000261783.3
|
ARG2
|
arginase 2 |
| chr16_-_66583701 | 0.10 |
ENST00000527800.1
ENST00000525974.1 ENST00000563369.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr4_+_5526883 | 0.10 |
ENST00000195455.2
|
C4orf6
|
chromosome 4 open reading frame 6 |
| chr6_-_31830655 | 0.10 |
ENST00000375631.4
|
NEU1
|
sialidase 1 (lysosomal sialidase) |
| chr2_+_32502952 | 0.10 |
ENST00000238831.4
|
YIPF4
|
Yip1 domain family, member 4 |
| chr3_-_123339418 | 0.10 |
ENST00000583087.1
|
MYLK
|
myosin light chain kinase |
| chr6_-_32095968 | 0.09 |
ENST00000375203.3
ENST00000375201.4 |
ATF6B
|
activating transcription factor 6 beta |
| chr18_+_61637159 | 0.09 |
ENST00000397985.2
ENST00000353706.2 ENST00000542677.1 ENST00000397988.3 ENST00000448851.1 |
SERPINB8
|
serpin peptidase inhibitor, clade B (ovalbumin), member 8 |
| chr6_-_76072719 | 0.09 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr1_+_160160283 | 0.09 |
ENST00000368079.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr12_+_7864021 | 0.09 |
ENST00000345088.2
|
DPPA3
|
developmental pluripotency associated 3 |
| chrX_-_119149501 | 0.09 |
ENST00000454625.1
|
GS1-421I3.2
|
GS1-421I3.2 |
| chr16_-_66584015 | 0.09 |
ENST00000545043.2
|
TK2
|
thymidine kinase 2, mitochondrial |
| chr4_+_5527117 | 0.08 |
ENST00000505296.1
|
C4orf6
|
chromosome 4 open reading frame 6 |
| chr11_-_66964638 | 0.08 |
ENST00000444002.2
|
AP001885.1
|
AP001885.1 |
| chr2_+_7073174 | 0.08 |
ENST00000416587.1
|
RNF144A
|
ring finger protein 144A |
| chr3_-_123339343 | 0.08 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr1_+_160160346 | 0.08 |
ENST00000368078.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr8_+_1993152 | 0.08 |
ENST00000262113.4
|
MYOM2
|
myomesin 2 |
| chr10_-_101380121 | 0.08 |
ENST00000370495.4
|
SLC25A28
|
solute carrier family 25 (mitochondrial iron transporter), member 28 |
| chr9_-_23779367 | 0.08 |
ENST00000440102.1
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr8_+_97597148 | 0.08 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr8_-_72274095 | 0.08 |
ENST00000303824.7
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr9_-_128246769 | 0.08 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr12_+_9980069 | 0.08 |
ENST00000354855.3
ENST00000324214.4 ENST00000279544.3 |
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1 |
| chr12_-_11150474 | 0.08 |
ENST00000538986.1
|
TAS2R20
|
taste receptor, type 2, member 20 |
| chrX_-_103499602 | 0.08 |
ENST00000372588.4
|
ESX1
|
ESX homeobox 1 |
| chr3_+_190105909 | 0.08 |
ENST00000456423.1
|
CLDN16
|
claudin 16 |
| chr12_+_14561422 | 0.08 |
ENST00000541056.1
|
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr19_+_55043977 | 0.07 |
ENST00000335056.3
|
KIR3DX1
|
killer cell immunoglobulin-like receptor, three domains, X1 |
| chr20_+_34129770 | 0.07 |
ENST00000348547.2
ENST00000357394.4 ENST00000447986.1 ENST00000279052.6 ENST00000416206.1 ENST00000411577.1 ENST00000413587.1 |
ERGIC3
|
ERGIC and golgi 3 |
| chr5_+_150404904 | 0.07 |
ENST00000521632.1
|
GPX3
|
glutathione peroxidase 3 (plasma) |
| chr4_+_119810134 | 0.07 |
ENST00000434046.2
|
SYNPO2
|
synaptopodin 2 |
| chr1_-_109935819 | 0.07 |
ENST00000538502.1
|
SORT1
|
sortilin 1 |
| chr1_-_19426149 | 0.07 |
ENST00000429347.2
|
UBR4
|
ubiquitin protein ligase E3 component n-recognin 4 |
| chr8_+_1993173 | 0.07 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr1_-_114429997 | 0.07 |
ENST00000471267.1
ENST00000393320.3 |
BCL2L15
|
BCL2-like 15 |
| chr1_-_167487758 | 0.06 |
ENST00000362089.5
|
CD247
|
CD247 molecule |
| chr18_-_3874271 | 0.06 |
ENST00000400149.3
ENST00000400155.1 ENST00000400150.3 |
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr18_-_3874247 | 0.06 |
ENST00000581699.1
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr10_+_90346519 | 0.06 |
ENST00000371939.3
|
LIPJ
|
lipase, family member J |
| chr11_+_92085262 | 0.06 |
ENST00000298047.6
ENST00000409404.2 ENST00000541502.1 |
FAT3
|
FAT atypical cadherin 3 |
| chr10_-_29923893 | 0.06 |
ENST00000355867.4
|
SVIL
|
supervillin |
| chr15_-_76304731 | 0.06 |
ENST00000394907.3
|
NRG4
|
neuregulin 4 |
| chr9_+_133539981 | 0.06 |
ENST00000253008.2
|
PRDM12
|
PR domain containing 12 |
| chr18_-_5396271 | 0.06 |
ENST00000579951.1
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr11_+_61197572 | 0.06 |
ENST00000542074.1
ENST00000534878.1 ENST00000537782.1 ENST00000543265.1 |
SDHAF2
|
succinate dehydrogenase complex assembly factor 2 |
| chr8_+_117963190 | 0.06 |
ENST00000427715.2
|
SLC30A8
|
solute carrier family 30 (zinc transporter), member 8 |
| chr3_+_108308513 | 0.06 |
ENST00000361582.3
|
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr2_+_202098203 | 0.06 |
ENST00000450491.1
ENST00000440732.1 ENST00000392258.3 |
CASP8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr1_-_72566613 | 0.06 |
ENST00000306821.3
|
NEGR1
|
neuronal growth regulator 1 |
| chr7_-_32338917 | 0.06 |
ENST00000396193.1
|
PDE1C
|
phosphodiesterase 1C, calmodulin-dependent 70kDa |
| chr11_+_12132117 | 0.06 |
ENST00000256194.4
|
MICAL2
|
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr7_+_36450169 | 0.06 |
ENST00000428612.1
|
ANLN
|
anillin, actin binding protein |
| chr1_-_35450897 | 0.06 |
ENST00000373337.3
|
ZMYM6NB
|
ZMYM6 neighbor |
| chrX_-_110507098 | 0.06 |
ENST00000541758.1
|
CAPN6
|
calpain 6 |
| chr8_+_24241969 | 0.06 |
ENST00000522298.1
|
ADAMDEC1
|
ADAM-like, decysin 1 |
| chr5_-_75919217 | 0.06 |
ENST00000504899.1
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr7_+_7811992 | 0.06 |
ENST00000406829.1
|
RPA3-AS1
|
RPA3 antisense RNA 1 |
| chr3_+_171561127 | 0.06 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chrX_+_134654540 | 0.05 |
ENST00000370752.4
|
DDX26B
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| chr12_-_82752565 | 0.05 |
ENST00000256151.7
|
CCDC59
|
coiled-coil domain containing 59 |
| chr1_+_8021954 | 0.05 |
ENST00000377491.1
ENST00000377488.1 |
PARK7
|
parkinson protein 7 |
| chr20_+_5731083 | 0.05 |
ENST00000445603.1
ENST00000442185.1 |
C20orf196
|
chromosome 20 open reading frame 196 |
| chr5_+_102594403 | 0.05 |
ENST00000319933.2
|
C5orf30
|
chromosome 5 open reading frame 30 |
| chr11_+_60681346 | 0.05 |
ENST00000227525.3
|
TMEM109
|
transmembrane protein 109 |
| chr10_+_97709725 | 0.05 |
ENST00000472454.2
|
RP11-248J23.6
|
Protein LOC100652732 |
| chr2_-_136288113 | 0.05 |
ENST00000401392.1
|
ZRANB3
|
zinc finger, RAN-binding domain containing 3 |
| chr9_+_75229616 | 0.05 |
ENST00000340019.3
|
TMC1
|
transmembrane channel-like 1 |
| chr13_+_50570019 | 0.05 |
ENST00000442421.1
|
TRIM13
|
tripartite motif containing 13 |
| chr8_+_24241789 | 0.05 |
ENST00000256412.4
ENST00000538205.1 |
ADAMDEC1
|
ADAM-like, decysin 1 |
| chr6_+_131894284 | 0.05 |
ENST00000368087.3
ENST00000356962.2 |
ARG1
|
arginase 1 |
| chr1_-_167487808 | 0.05 |
ENST00000392122.3
|
CD247
|
CD247 molecule |
| chr1_+_76540386 | 0.05 |
ENST00000328299.3
|
ST6GALNAC3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr13_-_88323218 | 0.05 |
ENST00000436290.2
ENST00000453832.2 ENST00000606590.1 |
MIR4500HG
|
MIR4500 host gene (non-protein coding) |
| chr12_-_10541575 | 0.04 |
ENST00000540818.1
|
KLRK1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr15_+_79166065 | 0.04 |
ENST00000559690.1
ENST00000559158.1 |
MORF4L1
|
mortality factor 4 like 1 |
| chr1_-_2461684 | 0.04 |
ENST00000378453.3
|
HES5
|
hes family bHLH transcription factor 5 |
| chr21_+_43619796 | 0.04 |
ENST00000398457.2
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr10_-_43892668 | 0.04 |
ENST00000544000.1
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr15_+_77712993 | 0.04 |
ENST00000336216.4
ENST00000381714.3 ENST00000558651.1 |
HMG20A
|
high mobility group 20A |
| chr4_+_113568207 | 0.03 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr13_-_45048386 | 0.03 |
ENST00000472477.1
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr9_-_15510287 | 0.03 |
ENST00000397519.2
|
PSIP1
|
PC4 and SFRS1 interacting protein 1 |
| chr10_+_32873190 | 0.03 |
ENST00000375025.4
|
C10orf68
|
Homo sapiens coiled-coil domain containing 7 (CCDC7), transcript variant 5, mRNA. |
| chr6_-_32784687 | 0.03 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr5_+_175490540 | 0.03 |
ENST00000515817.1
|
FAM153B
|
family with sequence similarity 153, member B |
| chr8_-_99057742 | 0.03 |
ENST00000521291.1
ENST00000396070.2 ENST00000523172.1 ENST00000287038.3 |
RPL30
|
ribosomal protein L30 |
| chr15_+_67813406 | 0.03 |
ENST00000342683.4
|
C15orf61
|
chromosome 15 open reading frame 61 |
| chr17_-_37934466 | 0.03 |
ENST00000583368.1
|
IKZF3
|
IKAROS family zinc finger 3 (Aiolos) |
| chr4_+_159442878 | 0.03 |
ENST00000307765.5
ENST00000423548.1 |
RXFP1
|
relaxin/insulin-like family peptide receptor 1 |
| chr3_+_93781728 | 0.02 |
ENST00000314622.4
|
NSUN3
|
NOP2/Sun domain family, member 3 |
| chr17_-_30228678 | 0.02 |
ENST00000261708.4
|
UTP6
|
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chr10_+_135204338 | 0.02 |
ENST00000468317.2
|
RP11-108K14.8
|
Mitochondrial GTPase 1 |
| chr3_+_114012819 | 0.02 |
ENST00000383671.3
|
TIGIT
|
T cell immunoreceptor with Ig and ITIM domains |
| chr4_+_159443024 | 0.02 |
ENST00000448688.2
|
RXFP1
|
relaxin/insulin-like family peptide receptor 1 |
| chr1_-_209792111 | 0.02 |
ENST00000455193.1
|
LAMB3
|
laminin, beta 3 |
| chrX_-_122756660 | 0.02 |
ENST00000441692.1
|
THOC2
|
THO complex 2 |
| chr6_+_136172820 | 0.02 |
ENST00000308191.6
|
PDE7B
|
phosphodiesterase 7B |
| chr22_+_41697520 | 0.02 |
ENST00000352645.4
|
ZC3H7B
|
zinc finger CCCH-type containing 7B |
| chrX_+_73164149 | 0.02 |
ENST00000602938.1
ENST00000602294.1 ENST00000602920.1 ENST00000602737.1 ENST00000602772.1 |
JPX
|
JPX transcript, XIST activator (non-protein coding) |
| chr16_+_69345243 | 0.02 |
ENST00000254950.11
|
VPS4A
|
vacuolar protein sorting 4 homolog A (S. cerevisiae) |
| chr15_-_77712429 | 0.02 |
ENST00000564328.1
ENST00000558305.1 |
PEAK1
|
pseudopodium-enriched atypical kinase 1 |
| chr1_+_44514040 | 0.02 |
ENST00000431800.1
ENST00000437643.1 |
RP5-1198O20.4
|
RP5-1198O20.4 |
| chr11_+_22694123 | 0.02 |
ENST00000534801.1
|
GAS2
|
growth arrest-specific 2 |
| chr1_+_46805832 | 0.02 |
ENST00000474844.1
|
NSUN4
|
NOP2/Sun domain family, member 4 |
| chr15_+_51669444 | 0.01 |
ENST00000396399.2
|
GLDN
|
gliomedin |
| chr17_-_42441204 | 0.01 |
ENST00000293443.7
|
FAM171A2
|
family with sequence similarity 171, member A2 |
| chr8_+_30244580 | 0.01 |
ENST00000523115.1
ENST00000519647.1 |
RBPMS
|
RNA binding protein with multiple splicing |
| chr5_+_140762268 | 0.01 |
ENST00000518325.1
|
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chr15_-_42500351 | 0.01 |
ENST00000348544.4
ENST00000318006.5 |
VPS39
|
vacuolar protein sorting 39 homolog (S. cerevisiae) |
| chr2_-_169746878 | 0.01 |
ENST00000282074.2
|
SPC25
|
SPC25, NDC80 kinetochore complex component |
| chr3_-_123512688 | 0.01 |
ENST00000475616.1
|
MYLK
|
myosin light chain kinase |
| chr20_+_56964169 | 0.01 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
Network of associatons between targets according to the STRING database.
First level regulatory network of MSX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 0.4 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.1 | 0.4 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.1 | 0.4 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.1 | 0.2 | GO:0072334 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.1 | 0.2 | GO:1902081 | regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.1 | 0.2 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.1 | 0.3 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.1 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.0 | 0.1 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.0 | 0.2 | GO:0042488 | positive regulation of odontogenesis of dentin-containing tooth(GO:0042488) positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.3 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:0038112 | interleukin-8-mediated signaling pathway(GO:0038112) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.1 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.2 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.1 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.0 | 0.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.3 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.1 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) |
| 0.0 | 0.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:1900111 | positive regulation of histone H3-K9 dimethylation(GO:1900111) |
| 0.0 | 0.1 | GO:0031106 | septin ring assembly(GO:0000921) septin ring organization(GO:0031106) |
| 0.0 | 0.1 | GO:1903181 | enzyme active site formation(GO:0018307) neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) detoxification of mercury ion(GO:0050787) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.0 | 0.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.0 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.0 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.0 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.0 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.3 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.3 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0039507 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.0 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:0043249 | erythrocyte maturation(GO:0043249) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.3 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.2 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.1 | 0.4 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.1 | 0.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.2 | GO:0004040 | amidase activity(GO:0004040) |
| 0.0 | 0.1 | GO:0047946 | glutamine N-acyltransferase activity(GO:0047946) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.1 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.2 | GO:0004918 | interleukin-8 receptor activity(GO:0004918) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.0 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.4 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.4 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.4 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |