Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
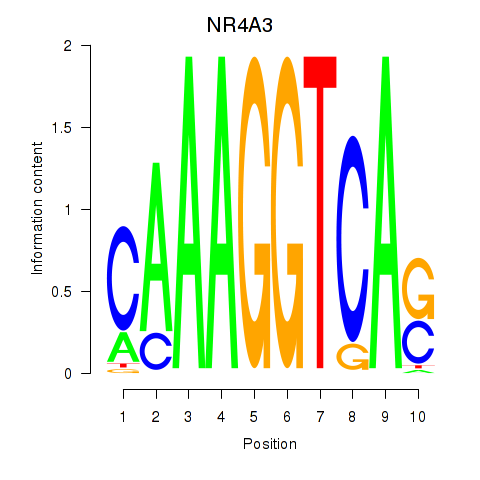
Results for NR4A3
Z-value: 0.45
Transcription factors associated with NR4A3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR4A3
|
ENSG00000119508.13 | nuclear receptor subfamily 4 group A member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR4A3 | hg19_v2_chr9_+_102584128_102584144 | -0.28 | 5.9e-01 | Click! |
Activity profile of NR4A3 motif
Sorted Z-values of NR4A3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_152552980 | 0.23 |
ENST00000368787.3
|
LCE3D
|
late cornified envelope 3D |
| chr11_-_73720276 | 0.21 |
ENST00000348534.4
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr8_-_131028782 | 0.20 |
ENST00000519020.1
|
FAM49B
|
family with sequence similarity 49, member B |
| chr11_+_73498973 | 0.20 |
ENST00000537007.1
|
MRPL48
|
mitochondrial ribosomal protein L48 |
| chr3_+_172034218 | 0.16 |
ENST00000366261.2
|
AC092964.1
|
Uncharacterized protein |
| chr22_+_46546406 | 0.16 |
ENST00000440343.1
ENST00000415785.1 |
PPARA
|
peroxisome proliferator-activated receptor alpha |
| chr9_-_127710292 | 0.15 |
ENST00000421514.1
|
GOLGA1
|
golgin A1 |
| chr2_+_88991162 | 0.15 |
ENST00000283646.4
|
RPIA
|
ribose 5-phosphate isomerase A |
| chr2_+_113763031 | 0.14 |
ENST00000259211.6
|
IL36A
|
interleukin 36, alpha |
| chr14_-_89878369 | 0.14 |
ENST00000553840.1
ENST00000556916.1 |
FOXN3
|
forkhead box N3 |
| chr20_-_23860373 | 0.13 |
ENST00000304710.4
|
CST5
|
cystatin D |
| chr14_-_104387888 | 0.13 |
ENST00000286953.3
|
C14orf2
|
chromosome 14 open reading frame 2 |
| chr10_-_30638090 | 0.13 |
ENST00000421701.1
ENST00000263063.4 |
MTPAP
|
mitochondrial poly(A) polymerase |
| chr12_-_95397442 | 0.12 |
ENST00000547157.1
ENST00000547986.1 ENST00000327772.2 |
NDUFA12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 |
| chr19_-_4535233 | 0.12 |
ENST00000381848.3
ENST00000588887.1 ENST00000586133.1 |
PLIN5
|
perilipin 5 |
| chr16_+_67563250 | 0.12 |
ENST00000566907.1
|
FAM65A
|
family with sequence similarity 65, member A |
| chr19_+_6372444 | 0.12 |
ENST00000245812.3
|
ALKBH7
|
alkB, alkylation repair homolog 7 (E. coli) |
| chr8_-_131028660 | 0.11 |
ENST00000401979.2
ENST00000517654.1 ENST00000522361.1 ENST00000518167.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr4_-_186732892 | 0.11 |
ENST00000451958.1
ENST00000439914.1 ENST00000428330.1 ENST00000429056.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr22_+_39493207 | 0.11 |
ENST00000348946.4
ENST00000442487.3 ENST00000421988.2 |
APOBEC3H
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3H |
| chr20_-_45530365 | 0.10 |
ENST00000414085.1
|
RP11-323C15.2
|
RP11-323C15.2 |
| chr22_+_31518938 | 0.10 |
ENST00000412985.1
ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr2_-_25194476 | 0.10 |
ENST00000534855.1
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr16_+_2880254 | 0.09 |
ENST00000570670.1
|
ZG16B
|
zymogen granule protein 16B |
| chr9_+_130890612 | 0.09 |
ENST00000443493.1
|
AL590708.2
|
AL590708.2 |
| chr8_-_131028869 | 0.09 |
ENST00000518283.1
ENST00000519110.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr2_+_9615179 | 0.09 |
ENST00000495797.1
|
IAH1
|
isoamyl acetate-hydrolyzing esterase 1 homolog (S. cerevisiae) |
| chr1_+_53392901 | 0.09 |
ENST00000371514.3
ENST00000528311.1 ENST00000371509.4 ENST00000407246.2 ENST00000371513.5 |
SCP2
|
sterol carrier protein 2 |
| chr19_+_35630628 | 0.09 |
ENST00000588715.1
ENST00000588607.1 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr11_-_68611721 | 0.09 |
ENST00000561996.1
|
CPT1A
|
carnitine palmitoyltransferase 1A (liver) |
| chrX_-_134049233 | 0.09 |
ENST00000370779.4
|
MOSPD1
|
motile sperm domain containing 1 |
| chr12_-_129308041 | 0.09 |
ENST00000376740.4
|
SLC15A4
|
solute carrier family 15 (oligopeptide transporter), member 4 |
| chr16_+_81040103 | 0.09 |
ENST00000305850.5
ENST00000299572.5 |
CENPN
|
centromere protein N |
| chr9_-_93727673 | 0.08 |
ENST00000427745.1
|
RP11-367F23.1
|
RP11-367F23.1 |
| chr16_+_29823427 | 0.08 |
ENST00000358758.7
ENST00000567659.1 ENST00000572820.1 |
PRRT2
|
proline-rich transmembrane protein 2 |
| chr11_-_118272610 | 0.08 |
ENST00000534438.1
|
RP11-770J1.5
|
Uncharacterized protein |
| chr15_+_66585555 | 0.08 |
ENST00000319194.5
ENST00000525134.2 ENST00000441424.2 |
DIS3L
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr22_+_39493268 | 0.08 |
ENST00000401756.1
|
APOBEC3H
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3H |
| chr19_+_589893 | 0.08 |
ENST00000251287.2
|
HCN2
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2 |
| chr11_+_118272328 | 0.08 |
ENST00000524422.1
|
ATP5L
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G |
| chr13_+_49822041 | 0.08 |
ENST00000538056.1
ENST00000251108.6 ENST00000444959.1 ENST00000429346.1 |
CDADC1
|
cytidine and dCMP deaminase domain containing 1 |
| chr21_-_27107283 | 0.08 |
ENST00000284971.3
ENST00000400099.1 |
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr5_-_1801408 | 0.08 |
ENST00000505818.1
|
MRPL36
|
mitochondrial ribosomal protein L36 |
| chr7_+_13141097 | 0.08 |
ENST00000411542.1
|
AC011288.2
|
AC011288.2 |
| chr2_+_151485403 | 0.08 |
ENST00000413247.1
ENST00000423428.1 ENST00000427615.1 ENST00000443811.1 |
AC104777.4
|
AC104777.4 |
| chr15_-_66790146 | 0.08 |
ENST00000316634.5
|
SNAPC5
|
small nuclear RNA activating complex, polypeptide 5, 19kDa |
| chr1_+_210502238 | 0.08 |
ENST00000545154.1
ENST00000537898.1 ENST00000391905.3 ENST00000545781.1 ENST00000261458.3 ENST00000308852.6 |
HHAT
|
hedgehog acyltransferase |
| chrX_-_48823165 | 0.08 |
ENST00000419374.1
|
KCND1
|
potassium voltage-gated channel, Shal-related subfamily, member 1 |
| chr1_-_11107280 | 0.07 |
ENST00000400897.3
ENST00000400898.3 |
MASP2
|
mannan-binding lectin serine peptidase 2 |
| chr14_-_104387831 | 0.07 |
ENST00000557040.1
ENST00000414262.2 ENST00000555030.1 ENST00000554713.1 ENST00000553430.1 |
C14orf2
|
chromosome 14 open reading frame 2 |
| chr16_+_83932684 | 0.07 |
ENST00000262430.4
|
MLYCD
|
malonyl-CoA decarboxylase |
| chr6_+_108616243 | 0.07 |
ENST00000421954.1
|
LACE1
|
lactation elevated 1 |
| chr11_-_71750865 | 0.07 |
ENST00000544129.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr12_+_123259063 | 0.07 |
ENST00000392441.4
ENST00000539171.1 |
CCDC62
|
coiled-coil domain containing 62 |
| chr3_-_156840776 | 0.07 |
ENST00000471357.1
|
LINC00880
|
long intergenic non-protein coding RNA 880 |
| chr14_+_37667118 | 0.07 |
ENST00000556615.1
ENST00000327441.7 ENST00000536774.1 |
MIPOL1
|
mirror-image polydactyly 1 |
| chr2_-_231989808 | 0.07 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr19_+_2977444 | 0.07 |
ENST00000246112.4
ENST00000453329.1 ENST00000482627.1 ENST00000452088.1 |
TLE6
|
transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) |
| chr1_+_46769303 | 0.07 |
ENST00000311672.5
|
UQCRH
|
ubiquinol-cytochrome c reductase hinge protein |
| chr1_-_219615984 | 0.07 |
ENST00000420762.1
|
RP11-95P13.1
|
RP11-95P13.1 |
| chr5_-_39424961 | 0.07 |
ENST00000503513.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr1_-_45253377 | 0.07 |
ENST00000372207.3
|
BEST4
|
bestrophin 4 |
| chr8_+_145065521 | 0.07 |
ENST00000534791.1
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr10_-_75351088 | 0.07 |
ENST00000451492.1
ENST00000413442.1 |
USP54
|
ubiquitin specific peptidase 54 |
| chr19_+_7011509 | 0.07 |
ENST00000377296.3
|
AC025278.1
|
Uncharacterized protein |
| chr21_-_46221684 | 0.07 |
ENST00000330942.5
|
UBE2G2
|
ubiquitin-conjugating enzyme E2G 2 |
| chr11_-_61560053 | 0.07 |
ENST00000537328.1
|
TMEM258
|
transmembrane protein 258 |
| chr12_+_117013656 | 0.07 |
ENST00000556529.1
|
MAP1LC3B2
|
microtubule-associated protein 1 light chain 3 beta 2 |
| chr10_-_14880002 | 0.07 |
ENST00000465530.1
|
CDNF
|
cerebral dopamine neurotrophic factor |
| chr13_-_95364389 | 0.06 |
ENST00000376945.2
|
SOX21
|
SRY (sex determining region Y)-box 21 |
| chr20_+_19738792 | 0.06 |
ENST00000412571.1
|
RP1-122P22.2
|
RP1-122P22.2 |
| chr9_+_35042205 | 0.06 |
ENST00000312292.5
ENST00000378745.3 |
C9orf131
|
chromosome 9 open reading frame 131 |
| chr1_-_169555779 | 0.06 |
ENST00000367797.3
ENST00000367796.3 |
F5
|
coagulation factor V (proaccelerin, labile factor) |
| chr2_+_238499812 | 0.06 |
ENST00000452801.1
|
AC104667.3
|
AC104667.3 |
| chr1_+_64059332 | 0.06 |
ENST00000540265.1
|
PGM1
|
phosphoglucomutase 1 |
| chr11_-_66206260 | 0.06 |
ENST00000329819.4
ENST00000310999.7 ENST00000430466.2 |
MRPL11
|
mitochondrial ribosomal protein L11 |
| chr20_-_60982330 | 0.06 |
ENST00000279101.5
|
CABLES2
|
Cdk5 and Abl enzyme substrate 2 |
| chr2_-_28113217 | 0.06 |
ENST00000444339.2
|
RBKS
|
ribokinase |
| chr1_+_16083098 | 0.06 |
ENST00000496928.2
ENST00000508310.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chrX_+_56259316 | 0.06 |
ENST00000468660.1
|
KLF8
|
Kruppel-like factor 8 |
| chr1_-_38218577 | 0.06 |
ENST00000540011.1
|
EPHA10
|
EPH receptor A10 |
| chr10_-_30637906 | 0.06 |
ENST00000417581.1
|
MTPAP
|
mitochondrial poly(A) polymerase |
| chr12_+_21679220 | 0.06 |
ENST00000256969.2
|
C12orf39
|
chromosome 12 open reading frame 39 |
| chr11_+_125496619 | 0.06 |
ENST00000532669.1
ENST00000278916.3 |
CHEK1
|
checkpoint kinase 1 |
| chr19_-_49176264 | 0.06 |
ENST00000270235.4
ENST00000596844.1 |
NTN5
|
netrin 5 |
| chr3_-_197300194 | 0.06 |
ENST00000358186.2
ENST00000431056.1 |
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr19_-_57967854 | 0.06 |
ENST00000321039.3
|
VN1R1
|
vomeronasal 1 receptor 1 |
| chr19_+_36239576 | 0.06 |
ENST00000587751.1
|
LIN37
|
lin-37 homolog (C. elegans) |
| chr1_-_12679171 | 0.06 |
ENST00000606790.1
|
RP11-474O21.5
|
RP11-474O21.5 |
| chr3_-_46000146 | 0.06 |
ENST00000438446.1
|
FYCO1
|
FYVE and coiled-coil domain containing 1 |
| chr8_-_101718991 | 0.06 |
ENST00000517990.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr7_+_23637763 | 0.06 |
ENST00000410069.1
|
CCDC126
|
coiled-coil domain containing 126 |
| chr17_-_79995553 | 0.06 |
ENST00000581584.1
ENST00000577712.1 ENST00000582900.1 ENST00000579155.1 ENST00000306869.2 |
DCXR
|
dicarbonyl/L-xylulose reductase |
| chr7_+_142498725 | 0.06 |
ENST00000466254.1
|
TRBC2
|
T cell receptor beta constant 2 |
| chr11_-_63993690 | 0.06 |
ENST00000394546.2
ENST00000541278.1 |
TRPT1
|
tRNA phosphotransferase 1 |
| chr11_+_3968573 | 0.06 |
ENST00000532990.1
|
STIM1
|
stromal interaction molecule 1 |
| chr15_-_55790515 | 0.06 |
ENST00000448430.2
ENST00000457155.2 |
DYX1C1
|
dyslexia susceptibility 1 candidate 1 |
| chr5_+_139055021 | 0.06 |
ENST00000502716.1
ENST00000503511.1 |
CXXC5
|
CXXC finger protein 5 |
| chr5_-_107703556 | 0.06 |
ENST00000496714.1
|
FBXL17
|
F-box and leucine-rich repeat protein 17 |
| chr20_-_32308028 | 0.06 |
ENST00000409299.3
ENST00000217398.3 ENST00000344022.3 |
PXMP4
|
peroxisomal membrane protein 4, 24kDa |
| chr4_-_48014931 | 0.06 |
ENST00000420489.2
ENST00000504722.1 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr19_+_19639670 | 0.06 |
ENST00000436027.5
|
YJEFN3
|
YjeF N-terminal domain containing 3 |
| chr3_-_9994021 | 0.06 |
ENST00000411976.2
ENST00000412055.1 |
PRRT3
|
proline-rich transmembrane protein 3 |
| chr10_+_81272287 | 0.05 |
ENST00000520547.2
|
EIF5AL1
|
eukaryotic translation initiation factor 5A-like 1 |
| chr14_-_81687575 | 0.05 |
ENST00000434192.2
|
GTF2A1
|
general transcription factor IIA, 1, 19/37kDa |
| chr1_+_171283331 | 0.05 |
ENST00000367749.3
|
FMO4
|
flavin containing monooxygenase 4 |
| chr3_+_179065474 | 0.05 |
ENST00000471841.1
ENST00000280653.7 |
MFN1
|
mitofusin 1 |
| chr7_+_45613958 | 0.05 |
ENST00000297323.7
|
ADCY1
|
adenylate cyclase 1 (brain) |
| chr12_+_109592477 | 0.05 |
ENST00000544726.1
|
ACACB
|
acetyl-CoA carboxylase beta |
| chr16_+_67840668 | 0.05 |
ENST00000415766.3
|
TSNAXIP1
|
translin-associated factor X interacting protein 1 |
| chr4_+_128886424 | 0.05 |
ENST00000398965.1
|
C4orf29
|
chromosome 4 open reading frame 29 |
| chr17_-_77967433 | 0.05 |
ENST00000571872.1
|
TBC1D16
|
TBC1 domain family, member 16 |
| chr1_+_14075903 | 0.05 |
ENST00000343137.4
ENST00000503842.1 ENST00000407521.3 ENST00000505823.1 |
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr12_+_70219052 | 0.05 |
ENST00000552032.2
ENST00000547771.2 |
MYRFL
|
myelin regulatory factor-like |
| chr17_+_61473104 | 0.05 |
ENST00000583016.1
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr3_-_139195350 | 0.05 |
ENST00000232217.2
|
RBP2
|
retinol binding protein 2, cellular |
| chr1_+_109234907 | 0.05 |
ENST00000370025.4
ENST00000370022.5 ENST00000370021.1 |
PRPF38B
|
pre-mRNA processing factor 38B |
| chr8_-_30584524 | 0.05 |
ENST00000521479.1
|
GSR
|
glutathione reductase |
| chr17_+_74536164 | 0.05 |
ENST00000586148.1
|
PRCD
|
progressive rod-cone degeneration |
| chr2_+_201242941 | 0.05 |
ENST00000449647.1
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr17_-_30669138 | 0.05 |
ENST00000225805.4
ENST00000577809.1 |
C17orf75
|
chromosome 17 open reading frame 75 |
| chr1_+_16083123 | 0.05 |
ENST00000510393.1
ENST00000430076.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr11_+_7595136 | 0.05 |
ENST00000529575.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr12_-_16035195 | 0.05 |
ENST00000535752.1
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr12_-_109219937 | 0.05 |
ENST00000546697.1
|
SSH1
|
slingshot protein phosphatase 1 |
| chr15_+_36887069 | 0.05 |
ENST00000566807.1
ENST00000567389.1 ENST00000562877.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr19_-_29218538 | 0.05 |
ENST00000592347.1
ENST00000586528.1 |
AC005307.3
|
AC005307.3 |
| chr1_-_151138323 | 0.05 |
ENST00000368908.5
|
LYSMD1
|
LysM, putative peptidoglycan-binding, domain containing 1 |
| chr5_+_149569520 | 0.05 |
ENST00000230671.2
ENST00000524041.1 |
SLC6A7
|
solute carrier family 6 (neurotransmitter transporter), member 7 |
| chrX_-_77225135 | 0.05 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr1_-_235116495 | 0.05 |
ENST00000549744.1
|
RP11-443B7.3
|
RP11-443B7.3 |
| chr17_-_2318731 | 0.05 |
ENST00000609667.1
|
AC006435.1
|
Uncharacterized protein |
| chr14_+_37667230 | 0.05 |
ENST00000556451.1
ENST00000556753.1 ENST00000396294.2 |
MIPOL1
|
mirror-image polydactyly 1 |
| chr7_+_121513143 | 0.05 |
ENST00000393386.2
|
PTPRZ1
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1 |
| chr2_+_26467825 | 0.05 |
ENST00000545822.1
ENST00000425035.1 |
HADHB
|
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), beta subunit |
| chr14_-_102976135 | 0.05 |
ENST00000560748.1
|
ANKRD9
|
ankyrin repeat domain 9 |
| chr11_-_66445219 | 0.05 |
ENST00000525754.1
ENST00000531969.1 ENST00000524637.1 ENST00000531036.2 ENST00000310046.4 |
RBM4B
|
RNA binding motif protein 4B |
| chr6_+_144185573 | 0.05 |
ENST00000237275.6
ENST00000539295.1 |
ZC2HC1B
|
zinc finger, C2HC-type containing 1B |
| chr19_-_41256207 | 0.05 |
ENST00000598485.2
ENST00000470681.1 ENST00000339153.3 ENST00000598729.1 |
C19orf54
|
chromosome 19 open reading frame 54 |
| chr20_-_18447667 | 0.05 |
ENST00000262547.5
ENST00000329494.5 ENST00000357236.4 |
DZANK1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr11_-_64900791 | 0.05 |
ENST00000531018.1
|
SYVN1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr3_-_9291063 | 0.04 |
ENST00000383836.3
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr22_-_39239987 | 0.04 |
ENST00000333039.2
|
NPTXR
|
neuronal pentraxin receptor |
| chr17_-_73178599 | 0.04 |
ENST00000578238.1
|
SUMO2
|
small ubiquitin-like modifier 2 |
| chr14_-_24911868 | 0.04 |
ENST00000554698.1
|
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr19_+_33622996 | 0.04 |
ENST00000592765.1
ENST00000361680.2 ENST00000355868.3 |
WDR88
|
WD repeat domain 88 |
| chr17_+_77893135 | 0.04 |
ENST00000574526.1
ENST00000572353.1 |
RP11-353N14.4
|
RP11-353N14.4 |
| chr17_-_39677971 | 0.04 |
ENST00000393976.2
|
KRT15
|
keratin 15 |
| chr14_-_30396948 | 0.04 |
ENST00000331968.5
|
PRKD1
|
protein kinase D1 |
| chr6_+_133561725 | 0.04 |
ENST00000452339.2
|
EYA4
|
eyes absent homolog 4 (Drosophila) |
| chr1_-_23521222 | 0.04 |
ENST00000374619.1
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr7_-_27169801 | 0.04 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr7_+_140396946 | 0.04 |
ENST00000476470.1
ENST00000471136.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr9_-_88896977 | 0.04 |
ENST00000311534.6
|
ISCA1
|
iron-sulfur cluster assembly 1 |
| chr3_-_46000064 | 0.04 |
ENST00000433878.1
|
FYCO1
|
FYVE and coiled-coil domain containing 1 |
| chr11_-_47736896 | 0.04 |
ENST00000525123.1
ENST00000528244.1 ENST00000532595.1 ENST00000529154.1 ENST00000530969.1 |
AGBL2
|
ATP/GTP binding protein-like 2 |
| chr9_-_122131696 | 0.04 |
ENST00000373964.2
ENST00000265922.3 |
BRINP1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 1 |
| chr1_-_154164534 | 0.04 |
ENST00000271850.7
ENST00000368530.2 |
TPM3
|
tropomyosin 3 |
| chr19_+_42363917 | 0.04 |
ENST00000598742.1
|
RPS19
|
ribosomal protein S19 |
| chr1_+_160175201 | 0.04 |
ENST00000368076.1
|
PEA15
|
phosphoprotein enriched in astrocytes 15 |
| chr4_+_159593418 | 0.04 |
ENST00000507475.1
ENST00000307738.5 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr15_-_23891175 | 0.04 |
ENST00000532292.1
|
MAGEL2
|
MAGE-like 2 |
| chr17_-_40976266 | 0.04 |
ENST00000593205.1
ENST00000361523.4 ENST00000590099.1 ENST00000438274.3 |
BECN1
|
beclin 1, autophagy related |
| chr19_+_11455900 | 0.04 |
ENST00000588790.1
|
CCDC159
|
coiled-coil domain containing 159 |
| chr7_-_68895857 | 0.04 |
ENST00000421513.1
|
RP11-3P22.2
|
RP11-3P22.2 |
| chr12_+_57854274 | 0.04 |
ENST00000528432.1
|
GLI1
|
GLI family zinc finger 1 |
| chr1_+_24118452 | 0.04 |
ENST00000421070.1
|
LYPLA2
|
lysophospholipase II |
| chr2_+_159313452 | 0.04 |
ENST00000389757.3
ENST00000389759.3 |
PKP4
|
plakophilin 4 |
| chr3_+_46449049 | 0.04 |
ENST00000357392.4
ENST00000400880.3 ENST00000433848.1 |
CCRL2
|
chemokine (C-C motif) receptor-like 2 |
| chr12_+_50898881 | 0.04 |
ENST00000301180.5
|
DIP2B
|
DIP2 disco-interacting protein 2 homolog B (Drosophila) |
| chr11_+_61015594 | 0.04 |
ENST00000451616.2
|
PGA5
|
pepsinogen 5, group I (pepsinogen A) |
| chr12_-_6715808 | 0.04 |
ENST00000545584.1
|
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr10_-_79397202 | 0.04 |
ENST00000372437.1
ENST00000372408.2 ENST00000372403.4 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr17_-_71258019 | 0.04 |
ENST00000344935.4
|
CPSF4L
|
cleavage and polyadenylation specific factor 4-like |
| chr8_+_99956662 | 0.04 |
ENST00000523368.1
ENST00000297565.4 ENST00000435298.2 |
OSR2
|
odd-skipped related transciption factor 2 |
| chr8_-_80942139 | 0.04 |
ENST00000521434.1
ENST00000519120.1 ENST00000520946.1 |
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr1_+_160175117 | 0.04 |
ENST00000360472.4
|
PEA15
|
phosphoprotein enriched in astrocytes 15 |
| chr13_-_114107839 | 0.04 |
ENST00000375418.3
|
ADPRHL1
|
ADP-ribosylhydrolase like 1 |
| chr5_+_175085033 | 0.04 |
ENST00000377291.2
|
HRH2
|
histamine receptor H2 |
| chr18_+_21719018 | 0.04 |
ENST00000585037.1
ENST00000415309.2 ENST00000399481.2 ENST00000577705.1 ENST00000327201.6 |
CABYR
|
calcium binding tyrosine-(Y)-phosphorylation regulated |
| chr19_-_18995029 | 0.04 |
ENST00000596048.1
|
CERS1
|
ceramide synthase 1 |
| chr3_+_29323043 | 0.04 |
ENST00000452462.1
ENST00000456853.1 |
RBMS3
|
RNA binding motif, single stranded interacting protein 3 |
| chr4_+_124317940 | 0.04 |
ENST00000505319.1
ENST00000339241.1 |
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chr5_-_160365601 | 0.04 |
ENST00000518580.1
ENST00000518301.1 |
RP11-109J4.1
|
RP11-109J4.1 |
| chr6_-_37467628 | 0.04 |
ENST00000373408.3
|
CCDC167
|
coiled-coil domain containing 167 |
| chr1_+_14075865 | 0.04 |
ENST00000413440.1
|
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr1_-_31661000 | 0.04 |
ENST00000263693.1
ENST00000398657.2 ENST00000526106.1 |
NKAIN1
|
Na+/K+ transporting ATPase interacting 1 |
| chr2_-_75796837 | 0.04 |
ENST00000233712.1
|
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr1_-_1356719 | 0.04 |
ENST00000520296.1
|
ANKRD65
|
ankyrin repeat domain 65 |
| chr19_+_17337473 | 0.04 |
ENST00000598068.1
|
OCEL1
|
occludin/ELL domain containing 1 |
| chr1_-_110052302 | 0.04 |
ENST00000369864.4
ENST00000369862.1 |
AMIGO1
|
adhesion molecule with Ig-like domain 1 |
| chrY_+_2870953 | 0.04 |
ENST00000444263.1
ENST00000425031.1 |
LINC00278
|
long intergenic non-protein coding RNA 278 |
| chr8_-_131028641 | 0.04 |
ENST00000523509.1
|
FAM49B
|
family with sequence similarity 49, member B |
| chr16_-_58328923 | 0.04 |
ENST00000567164.1
ENST00000219301.4 ENST00000569727.1 |
PRSS54
|
protease, serine, 54 |
| chr22_+_20104947 | 0.04 |
ENST00000402752.1
|
RANBP1
|
RAN binding protein 1 |
| chr7_-_38948774 | 0.04 |
ENST00000395969.2
ENST00000414632.1 ENST00000310301.4 |
VPS41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr10_-_116164450 | 0.04 |
ENST00000369271.3
|
AFAP1L2
|
actin filament associated protein 1-like 2 |
| chr15_-_52821247 | 0.04 |
ENST00000399231.3
ENST00000399233.2 |
MYO5A
|
myosin VA (heavy chain 12, myoxin) |
| chr12_+_119616447 | 0.04 |
ENST00000281938.2
|
HSPB8
|
heat shock 22kDa protein 8 |
| chr12_+_57828521 | 0.03 |
ENST00000309668.2
|
INHBC
|
inhibin, beta C |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR4A3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:2000568 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.1 | 0.2 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.2 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.0 | 0.2 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.0 | 0.1 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.0 | 0.1 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:0036146 | cellular response to mycotoxin(GO:0036146) |
| 0.0 | 0.1 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.0 | 0.0 | GO:1904717 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.1 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.0 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.0 | 0.1 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.0 | 0.0 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.0 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.1 | GO:0051643 | regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031585) endoplasmic reticulum localization(GO:0051643) |
| 0.0 | 0.0 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.0 | GO:0060032 | notochord regression(GO:0060032) |
| 0.0 | 0.1 | GO:0006388 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0010732 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.0 | 0.1 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.0 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.0 | 0.1 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.0 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.0 | GO:0035623 | renal glucose absorption(GO:0035623) |
| 0.0 | 0.0 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.0 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.0 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.2 | GO:0001884 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.1 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.1 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.0 | 0.1 | GO:0052856 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.0 | 0.1 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.0 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.1 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.0 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.0 | 0.0 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.0 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.0 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.0 | 0.0 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.1 | GO:0016679 | oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.0 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |