Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
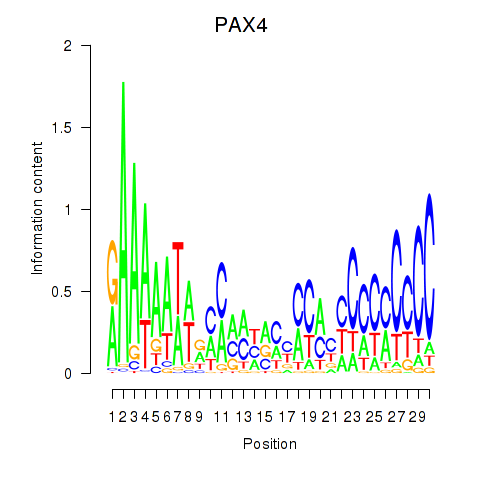
Results for PAX4
Z-value: 0.44
Transcription factors associated with PAX4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX4
|
ENSG00000106331.10 | paired box 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX4 | hg19_v2_chr7_-_127255982_127255982 | 0.47 | 3.5e-01 | Click! |
Activity profile of PAX4 motif
Sorted Z-values of PAX4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_2977444 | 0.34 |
ENST00000246112.4
ENST00000453329.1 ENST00000482627.1 ENST00000452088.1 |
TLE6
|
transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) |
| chr5_+_146614579 | 0.31 |
ENST00000541094.1
ENST00000398521.3 |
STK32A
|
serine/threonine kinase 32A |
| chr5_-_126409159 | 0.31 |
ENST00000607731.1
ENST00000535381.1 ENST00000296662.5 ENST00000509733.3 |
C5orf63
|
chromosome 5 open reading frame 63 |
| chr2_+_232316906 | 0.31 |
ENST00000370380.2
|
AC017104.2
|
Uncharacterized protein |
| chr19_-_17622269 | 0.30 |
ENST00000595116.1
|
CTD-3131K8.2
|
CTD-3131K8.2 |
| chr12_-_123717711 | 0.29 |
ENST00000537854.1
|
MPHOSPH9
|
M-phase phosphoprotein 9 |
| chr6_-_10694766 | 0.27 |
ENST00000460742.2
ENST00000259983.3 ENST00000379586.1 |
C6orf52
|
chromosome 6 open reading frame 52 |
| chr18_+_61254534 | 0.20 |
ENST00000269489.5
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13 |
| chrX_-_48827976 | 0.19 |
ENST00000218176.3
|
KCND1
|
potassium voltage-gated channel, Shal-related subfamily, member 1 |
| chr11_+_114549108 | 0.18 |
ENST00000389586.4
ENST00000375475.5 |
NXPE2
|
neurexophilin and PC-esterase domain family, member 2 |
| chr18_+_61254570 | 0.18 |
ENST00000344731.5
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr17_+_7253635 | 0.18 |
ENST00000571471.1
|
ACAP1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr1_+_33439268 | 0.18 |
ENST00000594612.1
|
FKSG48
|
FKSG48 |
| chr3_-_123339343 | 0.17 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr2_-_122042770 | 0.17 |
ENST00000263707.5
|
TFCP2L1
|
transcription factor CP2-like 1 |
| chr1_+_26644441 | 0.17 |
ENST00000374213.2
|
CD52
|
CD52 molecule |
| chr19_-_5624057 | 0.16 |
ENST00000590262.1
|
SAFB2
|
scaffold attachment factor B2 |
| chr16_-_28608424 | 0.16 |
ENST00000335715.4
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
| chr4_-_140544386 | 0.16 |
ENST00000561977.1
|
RP11-308D13.3
|
RP11-308D13.3 |
| chr9_+_92219919 | 0.16 |
ENST00000252506.6
ENST00000375769.1 |
GADD45G
|
growth arrest and DNA-damage-inducible, gamma |
| chr5_+_146614526 | 0.15 |
ENST00000397936.3
|
STK32A
|
serine/threonine kinase 32A |
| chr6_+_32605134 | 0.14 |
ENST00000343139.5
ENST00000395363.1 ENST00000496318.1 |
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chr4_+_165675269 | 0.14 |
ENST00000507311.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr2_+_99797636 | 0.14 |
ENST00000409145.1
|
MRPL30
|
mitochondrial ribosomal protein L30 |
| chr3_+_119298523 | 0.13 |
ENST00000357003.3
|
ADPRH
|
ADP-ribosylarginine hydrolase |
| chr6_+_133135580 | 0.13 |
ENST00000230050.3
|
RPS12
|
ribosomal protein S12 |
| chr18_-_53255766 | 0.13 |
ENST00000566286.1
ENST00000564999.1 ENST00000566279.1 ENST00000354452.3 ENST00000356073.4 |
TCF4
|
transcription factor 4 |
| chr8_+_67344710 | 0.13 |
ENST00000379385.4
ENST00000396623.3 ENST00000415254.1 |
ADHFE1
|
alcohol dehydrogenase, iron containing, 1 |
| chr12_+_56546363 | 0.13 |
ENST00000551834.1
ENST00000552568.1 |
MYL6B
|
myosin, light chain 6B, alkali, smooth muscle and non-muscle |
| chr14_+_78227105 | 0.12 |
ENST00000439131.2
ENST00000355883.3 ENST00000557011.1 ENST00000556047.1 |
C14orf178
|
chromosome 14 open reading frame 178 |
| chr5_+_150406527 | 0.12 |
ENST00000520059.1
|
GPX3
|
glutathione peroxidase 3 (plasma) |
| chr17_-_46799872 | 0.12 |
ENST00000290294.3
|
PRAC1
|
prostate cancer susceptibility candidate 1 |
| chr2_-_191184571 | 0.12 |
ENST00000392332.3
|
HIBCH
|
3-hydroxyisobutyryl-CoA hydrolase |
| chr10_-_70092635 | 0.12 |
ENST00000309049.4
|
PBLD
|
phenazine biosynthesis-like protein domain containing |
| chr1_+_244816237 | 0.11 |
ENST00000302550.11
|
DESI2
|
desumoylating isopeptidase 2 |
| chr1_-_152061537 | 0.11 |
ENST00000368806.1
|
TCHHL1
|
trichohyalin-like 1 |
| chr16_-_67224002 | 0.11 |
ENST00000563889.1
ENST00000564418.1 ENST00000545725.2 ENST00000314586.6 |
EXOC3L1
|
exocyst complex component 3-like 1 |
| chr19_+_10959043 | 0.11 |
ENST00000397820.4
|
C19orf38
|
chromosome 19 open reading frame 38 |
| chr3_-_123339418 | 0.11 |
ENST00000583087.1
|
MYLK
|
myosin light chain kinase |
| chr10_-_70092671 | 0.11 |
ENST00000358769.2
ENST00000432941.1 ENST00000495025.2 |
PBLD
|
phenazine biosynthesis-like protein domain containing |
| chr7_+_26331678 | 0.11 |
ENST00000446848.2
|
SNX10
|
sorting nexin 10 |
| chr12_+_116997186 | 0.11 |
ENST00000306985.4
|
MAP1LC3B2
|
microtubule-associated protein 1 light chain 3 beta 2 |
| chr1_+_101702417 | 0.11 |
ENST00000305352.6
|
S1PR1
|
sphingosine-1-phosphate receptor 1 |
| chr8_+_143781883 | 0.11 |
ENST00000522591.1
|
LY6K
|
lymphocyte antigen 6 complex, locus K |
| chr19_+_11708259 | 0.11 |
ENST00000587939.1
ENST00000588174.1 |
ZNF627
|
zinc finger protein 627 |
| chr6_-_113754604 | 0.10 |
ENST00000421737.1
|
RP1-124C6.1
|
RP1-124C6.1 |
| chr1_-_231376836 | 0.10 |
ENST00000451322.1
|
C1orf131
|
chromosome 1 open reading frame 131 |
| chr18_-_53089538 | 0.10 |
ENST00000566777.1
|
TCF4
|
transcription factor 4 |
| chr18_+_61254221 | 0.10 |
ENST00000431153.1
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr18_-_53089723 | 0.10 |
ENST00000561992.1
ENST00000562512.2 |
TCF4
|
transcription factor 4 |
| chr9_+_134103496 | 0.10 |
ENST00000498010.1
ENST00000476004.1 ENST00000528406.1 |
NUP214
|
nucleoporin 214kDa |
| chr4_-_113207048 | 0.10 |
ENST00000361717.3
|
TIFA
|
TRAF-interacting protein with forkhead-associated domain |
| chr17_+_7254184 | 0.10 |
ENST00000575415.1
|
ACAP1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr7_+_80267973 | 0.10 |
ENST00000394788.3
ENST00000447544.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr11_-_124622134 | 0.10 |
ENST00000326621.5
|
VSIG2
|
V-set and immunoglobulin domain containing 2 |
| chr4_-_176733897 | 0.10 |
ENST00000393658.2
|
GPM6A
|
glycoprotein M6A |
| chr4_-_76439596 | 0.09 |
ENST00000451788.1
ENST00000512706.1 |
RCHY1
|
ring finger and CHY zinc finger domain containing 1, E3 ubiquitin protein ligase |
| chr10_-_14920599 | 0.09 |
ENST00000609399.1
|
RP11-398C13.6
|
RP11-398C13.6 |
| chr9_-_100000957 | 0.09 |
ENST00000366109.2
ENST00000607322.1 |
RP11-498P14.5
|
RP11-498P14.5 |
| chr19_+_11708229 | 0.09 |
ENST00000361113.5
|
ZNF627
|
zinc finger protein 627 |
| chr4_+_30723003 | 0.09 |
ENST00000543491.1
|
PCDH7
|
protocadherin 7 |
| chr10_-_25012115 | 0.09 |
ENST00000446003.1
|
ARHGAP21
|
Rho GTPase activating protein 21 |
| chr17_+_1674982 | 0.09 |
ENST00000572048.1
ENST00000573763.1 |
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr15_+_69373210 | 0.09 |
ENST00000435479.1
ENST00000559870.1 |
LINC00277
RP11-809H16.5
|
long intergenic non-protein coding RNA 277 RP11-809H16.5 |
| chrX_-_102943022 | 0.09 |
ENST00000433176.2
|
MORF4L2
|
mortality factor 4 like 2 |
| chr12_+_110718428 | 0.09 |
ENST00000552636.1
|
ATP2A2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
| chr4_+_140586922 | 0.09 |
ENST00000265498.1
ENST00000506797.1 |
MGST2
|
microsomal glutathione S-transferase 2 |
| chr2_-_85839146 | 0.09 |
ENST00000306336.5
ENST00000409734.3 |
C2orf68
|
chromosome 2 open reading frame 68 |
| chr6_+_32605195 | 0.08 |
ENST00000374949.2
|
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chrX_-_101410762 | 0.08 |
ENST00000543160.1
ENST00000333643.3 |
BEX5
|
brain expressed, X-linked 5 |
| chr12_+_123717458 | 0.08 |
ENST00000253233.1
|
C12orf65
|
chromosome 12 open reading frame 65 |
| chr14_-_78227476 | 0.08 |
ENST00000554775.1
ENST00000555761.1 ENST00000554324.1 ENST00000261531.7 |
SNW1
|
SNW domain containing 1 |
| chr1_-_1677358 | 0.08 |
ENST00000355439.2
ENST00000400924.1 ENST00000246421.4 |
SLC35E2
|
solute carrier family 35, member E2 |
| chr4_-_169931393 | 0.08 |
ENST00000504480.1
ENST00000306193.3 |
CBR4
|
carbonyl reductase 4 |
| chr1_+_145525015 | 0.08 |
ENST00000539363.1
ENST00000538811.1 |
ITGA10
|
integrin, alpha 10 |
| chr7_+_134916731 | 0.08 |
ENST00000275764.3
|
STRA8
|
stimulated by retinoic acid 8 |
| chr19_+_55141861 | 0.08 |
ENST00000396327.3
ENST00000324602.7 ENST00000434867.2 |
LILRB1
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 1 |
| chr1_+_244816371 | 0.08 |
ENST00000263831.7
|
DESI2
|
desumoylating isopeptidase 2 |
| chr13_-_25496926 | 0.08 |
ENST00000545981.1
ENST00000381884.4 |
CENPJ
|
centromere protein J |
| chr2_+_99758161 | 0.08 |
ENST00000409684.1
|
C2ORF15
|
Uncharacterized protein C2orf15 |
| chr3_-_142166904 | 0.07 |
ENST00000264951.4
|
XRN1
|
5'-3' exoribonuclease 1 |
| chr18_-_53255379 | 0.07 |
ENST00000565908.2
|
TCF4
|
transcription factor 4 |
| chr12_-_95044309 | 0.07 |
ENST00000261226.4
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr15_+_63340647 | 0.07 |
ENST00000404484.4
|
TPM1
|
tropomyosin 1 (alpha) |
| chr19_-_51961702 | 0.07 |
ENST00000430817.1
ENST00000321424.3 ENST00000340550.5 |
SIGLEC8
|
sialic acid binding Ig-like lectin 8 |
| chr4_+_174292058 | 0.07 |
ENST00000296504.3
|
SAP30
|
Sin3A-associated protein, 30kDa |
| chrX_-_131623982 | 0.07 |
ENST00000370844.1
|
MBNL3
|
muscleblind-like splicing regulator 3 |
| chr8_-_101963482 | 0.07 |
ENST00000419477.2
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr16_-_28608364 | 0.07 |
ENST00000533150.1
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
| chr4_+_95129061 | 0.07 |
ENST00000354268.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr4_+_95128996 | 0.07 |
ENST00000457823.2
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr12_+_118573663 | 0.07 |
ENST00000261313.2
|
PEBP1
|
phosphatidylethanolamine binding protein 1 |
| chr3_-_142166846 | 0.07 |
ENST00000463916.1
ENST00000544157.1 |
XRN1
|
5'-3' exoribonuclease 1 |
| chr4_+_170541678 | 0.07 |
ENST00000360642.3
ENST00000512813.1 |
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr4_+_47487285 | 0.06 |
ENST00000273859.3
ENST00000504445.1 |
ATP10D
|
ATPase, class V, type 10D |
| chr1_-_85742773 | 0.06 |
ENST00000370580.1
|
BCL10
|
B-cell CLL/lymphoma 10 |
| chr4_-_492891 | 0.06 |
ENST00000338977.5
ENST00000511833.2 |
ZNF721
|
zinc finger protein 721 |
| chr4_+_170541835 | 0.06 |
ENST00000504131.2
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr7_+_134233840 | 0.06 |
ENST00000457545.2
|
AKR1B15
|
aldo-keto reductase family 1, member B15 |
| chr12_-_66035922 | 0.06 |
ENST00000546198.1
ENST00000535315.1 ENST00000537298.1 |
RP11-230G5.2
|
RP11-230G5.2 |
| chrX_-_131547596 | 0.06 |
ENST00000538204.1
ENST00000370849.3 |
MBNL3
|
muscleblind-like splicing regulator 3 |
| chr16_-_49890016 | 0.06 |
ENST00000563137.2
|
ZNF423
|
zinc finger protein 423 |
| chr1_-_240775447 | 0.06 |
ENST00000318160.4
|
GREM2
|
gremlin 2, DAN family BMP antagonist |
| chr19_+_9361606 | 0.06 |
ENST00000456448.1
|
OR7E24
|
olfactory receptor, family 7, subfamily E, member 24 |
| chr4_+_170541660 | 0.06 |
ENST00000513761.1
ENST00000347613.4 |
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr16_-_79804394 | 0.05 |
ENST00000567993.1
|
RP11-345M22.2
|
RP11-345M22.2 |
| chr18_-_3013307 | 0.05 |
ENST00000584294.1
|
LPIN2
|
lipin 2 |
| chr3_+_69134124 | 0.05 |
ENST00000478935.1
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr21_-_33651324 | 0.05 |
ENST00000290130.3
|
MIS18A
|
MIS18 kinetochore protein A |
| chr12_+_16064258 | 0.05 |
ENST00000524480.1
ENST00000531803.1 ENST00000532964.1 |
DERA
|
deoxyribose-phosphate aldolase (putative) |
| chr11_+_20385327 | 0.05 |
ENST00000451739.2
ENST00000532505.1 |
HTATIP2
|
HIV-1 Tat interactive protein 2, 30kDa |
| chr6_-_90062543 | 0.05 |
ENST00000435041.2
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1 |
| chr1_+_231376941 | 0.05 |
ENST00000436239.1
ENST00000366647.4 ENST00000366646.3 ENST00000416000.1 |
GNPAT
|
glyceronephosphate O-acyltransferase |
| chr12_-_11091862 | 0.05 |
ENST00000537503.1
|
TAS2R14
|
taste receptor, type 2, member 14 |
| chr3_+_107318157 | 0.05 |
ENST00000406780.1
|
BBX
|
bobby sox homolog (Drosophila) |
| chr12_+_16064106 | 0.04 |
ENST00000428559.2
|
DERA
|
deoxyribose-phosphate aldolase (putative) |
| chr3_+_27754397 | 0.04 |
ENST00000606069.1
|
RP11-222K16.2
|
RP11-222K16.2 |
| chr15_+_63340553 | 0.04 |
ENST00000334895.5
|
TPM1
|
tropomyosin 1 (alpha) |
| chr4_+_39184024 | 0.04 |
ENST00000399820.3
ENST00000509560.1 ENST00000512112.1 ENST00000288634.7 ENST00000506503.1 |
WDR19
|
WD repeat domain 19 |
| chr6_+_32407619 | 0.04 |
ENST00000395388.2
ENST00000374982.5 |
HLA-DRA
|
major histocompatibility complex, class II, DR alpha |
| chrX_+_135067576 | 0.04 |
ENST00000370701.1
ENST00000370698.3 ENST00000370695.4 |
SLC9A6
|
solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6 |
| chr19_+_55141948 | 0.04 |
ENST00000396332.4
ENST00000427581.2 |
LILRB1
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 1 |
| chr5_+_119799927 | 0.04 |
ENST00000407149.2
ENST00000379551.2 |
PRR16
|
proline rich 16 |
| chr2_+_201173667 | 0.04 |
ENST00000409755.3
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chrX_-_131547625 | 0.04 |
ENST00000394311.2
|
MBNL3
|
muscleblind-like splicing regulator 3 |
| chr5_+_118668846 | 0.04 |
ENST00000513374.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr19_-_52552051 | 0.04 |
ENST00000221315.5
|
ZNF432
|
zinc finger protein 432 |
| chr12_+_31812121 | 0.04 |
ENST00000395763.3
|
METTL20
|
methyltransferase like 20 |
| chr5_-_175395283 | 0.04 |
ENST00000513482.1
ENST00000265097.4 |
THOC3
|
THO complex 3 |
| chrX_-_31284974 | 0.04 |
ENST00000378702.4
|
DMD
|
dystrophin |
| chr5_+_173473662 | 0.04 |
ENST00000519717.1
|
NSG2
|
Neuron-specific protein family member 2 |
| chr18_+_29027696 | 0.04 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr8_-_74205851 | 0.04 |
ENST00000396467.1
|
RPL7
|
ribosomal protein L7 |
| chr19_-_53193731 | 0.04 |
ENST00000598536.1
ENST00000594682.2 ENST00000601257.1 |
ZNF83
|
zinc finger protein 83 |
| chr2_+_190526111 | 0.04 |
ENST00000607062.1
ENST00000260952.4 ENST00000425590.1 ENST00000607535.1 ENST00000420250.1 ENST00000606910.1 ENST00000607690.1 ENST00000607829.1 |
ASNSD1
|
asparagine synthetase domain containing 1 |
| chr14_+_76072096 | 0.04 |
ENST00000555058.1
|
FLVCR2
|
feline leukemia virus subgroup C cellular receptor family, member 2 |
| chr11_-_111741994 | 0.04 |
ENST00000398006.2
|
ALG9
|
ALG9, alpha-1,2-mannosyltransferase |
| chr7_+_133812052 | 0.04 |
ENST00000285928.2
|
LRGUK
|
leucine-rich repeats and guanylate kinase domain containing |
| chr8_-_38853990 | 0.04 |
ENST00000456845.2
ENST00000397070.2 ENST00000517872.1 ENST00000412303.1 ENST00000456397.2 |
TM2D2
|
TM2 domain containing 2 |
| chr5_-_125930877 | 0.04 |
ENST00000510111.2
ENST00000509270.1 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr22_-_31688381 | 0.04 |
ENST00000487265.2
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr1_+_210001309 | 0.04 |
ENST00000491415.2
|
DIEXF
|
digestive organ expansion factor homolog (zebrafish) |
| chr13_+_28712614 | 0.03 |
ENST00000380958.3
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chrX_-_19533379 | 0.03 |
ENST00000338883.4
|
MAP3K15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr7_-_130080818 | 0.03 |
ENST00000343969.5
ENST00000541543.1 ENST00000489512.1 |
CEP41
|
centrosomal protein 41kDa |
| chr15_-_72514866 | 0.03 |
ENST00000562997.1
|
PKM
|
pyruvate kinase, muscle |
| chr7_-_16872932 | 0.03 |
ENST00000419572.2
ENST00000412973.1 |
AGR2
|
anterior gradient 2 |
| chr16_+_53088885 | 0.03 |
ENST00000566029.1
ENST00000447540.1 |
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr2_+_47630108 | 0.03 |
ENST00000233146.2
ENST00000454849.1 ENST00000543555.1 |
MSH2
|
mutS homolog 2 |
| chr17_-_74049720 | 0.03 |
ENST00000602720.1
|
SRP68
|
signal recognition particle 68kDa |
| chr2_-_208489707 | 0.03 |
ENST00000448007.2
ENST00000432416.1 ENST00000411432.1 |
METTL21A
|
methyltransferase like 21A |
| chr2_-_10830093 | 0.03 |
ENST00000381685.5
ENST00000345985.3 ENST00000542668.1 ENST00000538384.1 |
NOL10
|
nucleolar protein 10 |
| chr5_-_54988448 | 0.03 |
ENST00000503817.1
ENST00000512595.1 |
SLC38A9
|
solute carrier family 38, member 9 |
| chr6_+_134274354 | 0.03 |
ENST00000367869.1
|
TBPL1
|
TBP-like 1 |
| chr8_-_101963677 | 0.03 |
ENST00000395956.3
ENST00000395953.2 |
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr1_+_162039558 | 0.03 |
ENST00000530878.1
ENST00000361897.5 |
NOS1AP
|
nitric oxide synthase 1 (neuronal) adaptor protein |
| chr11_+_46740730 | 0.03 |
ENST00000311907.5
ENST00000530231.1 ENST00000442468.1 |
F2
|
coagulation factor II (thrombin) |
| chr2_-_20251744 | 0.03 |
ENST00000175091.4
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha |
| chr7_-_96654133 | 0.03 |
ENST00000486603.2
ENST00000222598.4 |
DLX5
|
distal-less homeobox 5 |
| chr11_+_1874200 | 0.03 |
ENST00000311604.3
|
LSP1
|
lymphocyte-specific protein 1 |
| chr2_+_15731289 | 0.03 |
ENST00000381341.2
|
DDX1
|
DEAD (Asp-Glu-Ala-Asp) box helicase 1 |
| chr11_+_57529234 | 0.03 |
ENST00000360682.6
ENST00000361796.4 ENST00000529526.1 ENST00000426142.2 ENST00000399050.4 ENST00000361391.6 ENST00000361332.4 ENST00000532463.1 ENST00000529986.1 ENST00000358694.6 ENST00000532787.1 ENST00000533667.1 ENST00000532649.1 ENST00000528621.1 ENST00000530748.1 ENST00000428599.2 ENST00000527467.1 ENST00000528232.1 ENST00000531014.1 ENST00000526772.1 ENST00000529873.1 ENST00000525902.1 ENST00000532844.1 ENST00000526357.1 ENST00000530094.1 ENST00000415361.2 ENST00000532245.1 ENST00000534579.1 ENST00000526938.1 |
CTNND1
|
catenin (cadherin-associated protein), delta 1 |
| chr2_+_231860830 | 0.03 |
ENST00000424440.1
ENST00000452881.1 ENST00000433428.2 ENST00000455816.1 ENST00000440792.1 ENST00000423134.1 |
SPATA3
|
spermatogenesis associated 3 |
| chr14_-_50559361 | 0.03 |
ENST00000305273.1
|
C14orf183
|
chromosome 14 open reading frame 183 |
| chr21_+_43934633 | 0.03 |
ENST00000398343.2
|
SLC37A1
|
solute carrier family 37 (glucose-6-phosphate transporter), member 1 |
| chrX_-_102942961 | 0.03 |
ENST00000434230.1
ENST00000418819.1 ENST00000360458.1 |
MORF4L2
|
mortality factor 4 like 2 |
| chr11_+_20385231 | 0.03 |
ENST00000530266.1
ENST00000421577.2 ENST00000443524.2 ENST00000419348.2 |
HTATIP2
|
HIV-1 Tat interactive protein 2, 30kDa |
| chr17_-_33390667 | 0.03 |
ENST00000378516.2
ENST00000268850.7 ENST00000394597.2 |
RFFL
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr11_+_134144139 | 0.03 |
ENST00000389887.5
|
GLB1L3
|
galactosidase, beta 1-like 3 |
| chr1_+_90308981 | 0.03 |
ENST00000527156.1
|
LRRC8D
|
leucine rich repeat containing 8 family, member D |
| chr9_+_100000717 | 0.03 |
ENST00000375205.2
ENST00000357054.1 ENST00000395220.1 ENST00000375202.2 ENST00000411667.2 |
CCDC180
|
coiled-coil domain containing 180 |
| chr4_-_84030996 | 0.03 |
ENST00000411416.2
|
PLAC8
|
placenta-specific 8 |
| chr7_-_86595190 | 0.02 |
ENST00000398276.2
ENST00000416314.1 ENST00000425689.1 |
KIAA1324L
|
KIAA1324-like |
| chr2_-_217560248 | 0.02 |
ENST00000233813.4
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr8_-_116680833 | 0.02 |
ENST00000220888.5
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr5_-_142782862 | 0.02 |
ENST00000415690.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr8_-_13134045 | 0.02 |
ENST00000512044.2
|
DLC1
|
deleted in liver cancer 1 |
| chr16_-_28621312 | 0.02 |
ENST00000314752.7
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr2_+_216176761 | 0.02 |
ENST00000540518.1
ENST00000435675.1 |
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase |
| chr18_-_32957260 | 0.02 |
ENST00000587422.1
ENST00000306346.1 ENST00000589332.1 ENST00000586687.1 ENST00000585522.1 |
ZNF396
|
zinc finger protein 396 |
| chr1_-_151032040 | 0.02 |
ENST00000540998.1
ENST00000357235.5 |
CDC42SE1
|
CDC42 small effector 1 |
| chr11_-_124622083 | 0.02 |
ENST00000403470.1
|
VSIG2
|
V-set and immunoglobulin domain containing 2 |
| chr15_-_82641706 | 0.02 |
ENST00000439287.4
|
GOLGA6L10
|
golgin A6 family-like 10 |
| chr11_-_93276582 | 0.02 |
ENST00000298966.2
|
SMCO4
|
single-pass membrane protein with coiled-coil domains 4 |
| chr16_-_79804435 | 0.02 |
ENST00000562921.1
ENST00000566729.1 |
RP11-345M22.2
|
RP11-345M22.2 |
| chr4_-_169931231 | 0.02 |
ENST00000504561.1
|
CBR4
|
carbonyl reductase 4 |
| chr1_+_162601774 | 0.02 |
ENST00000415555.1
|
DDR2
|
discoidin domain receptor tyrosine kinase 2 |
| chr2_-_208489275 | 0.02 |
ENST00000272839.3
ENST00000426075.1 |
METTL21A
|
methyltransferase like 21A |
| chr3_+_46283863 | 0.02 |
ENST00000545097.1
ENST00000541018.1 |
CCR3
|
chemokine (C-C motif) receptor 3 |
| chr17_-_26697304 | 0.02 |
ENST00000536498.1
|
VTN
|
vitronectin |
| chr2_+_99797542 | 0.02 |
ENST00000338148.3
ENST00000512183.2 |
MRPL30
C2orf15
|
mitochondrial ribosomal protein L30 chromosome 2 open reading frame 15 |
| chr11_-_96239990 | 0.02 |
ENST00000511243.2
|
JRKL-AS1
|
JRKL antisense RNA 1 |
| chr4_-_171011323 | 0.02 |
ENST00000509167.1
ENST00000353187.2 ENST00000507375.1 ENST00000515480.1 |
AADAT
|
aminoadipate aminotransferase |
| chr19_+_14544099 | 0.02 |
ENST00000242783.6
ENST00000586557.1 ENST00000590097.1 |
PKN1
|
protein kinase N1 |
| chr7_-_23053719 | 0.02 |
ENST00000432176.2
ENST00000440481.1 |
FAM126A
|
family with sequence similarity 126, member A |
| chr13_-_46543805 | 0.02 |
ENST00000378921.2
|
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr1_+_145524891 | 0.02 |
ENST00000369304.3
|
ITGA10
|
integrin, alpha 10 |
| chr9_-_20621834 | 0.02 |
ENST00000429426.2
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chrX_+_106163626 | 0.02 |
ENST00000336803.1
|
CLDN2
|
claudin 2 |
| chr5_-_20575959 | 0.02 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1902173 | negative regulation of keratinocyte apoptotic process(GO:1902173) |
| 0.1 | 0.2 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.1 | GO:0015993 | molecular hydrogen transport(GO:0015993) |
| 0.0 | 0.1 | GO:0035548 | gamma-delta T cell activation involved in immune response(GO:0002290) negative regulation of interferon-beta secretion(GO:0035548) regulation of gamma-delta T cell activation involved in immune response(GO:2001191) positive regulation of gamma-delta T cell activation involved in immune response(GO:2001193) |
| 0.0 | 0.3 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.1 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.3 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.2 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:1903515 | calcium ion transport from cytosol to endoplasmic reticulum(GO:1903515) |
| 0.0 | 0.1 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 0.1 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.1 | GO:0032765 | lymphotoxin A production(GO:0032641) positive regulation of mast cell cytokine production(GO:0032765) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.0 | 0.1 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.0 | GO:0031455 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.2 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.0 | 0.0 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.0 | 0.0 | GO:1905033 | positive regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903762) positive regulation of ventricular cardiac muscle cell action potential(GO:1903947) positive regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905026) positive regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905033) |
| 0.0 | 0.0 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.0 | GO:0060166 | olfactory pit development(GO:0060166) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.1 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.0 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030109 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) HLA-B specific inhibitory MHC class I receptor activity(GO:0030109) inhibitory MHC class I receptor activity(GO:0032396) |
| 0.0 | 0.1 | GO:0003955 | NAD(P)H dehydrogenase (quinone) activity(GO:0003955) 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 0.1 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.0 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0086039 | lutropin-choriogonadotropic hormone receptor binding(GO:0031775) calcium-transporting ATPase activity involved in regulation of cardiac muscle cell membrane potential(GO:0086039) |
| 0.0 | 0.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.4 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.3 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |