Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
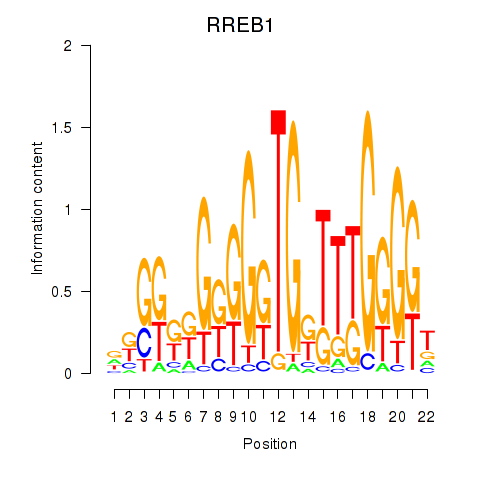
Results for RREB1
Z-value: 0.45
Transcription factors associated with RREB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RREB1
|
ENSG00000124782.15 | ras responsive element binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RREB1 | hg19_v2_chr6_+_7107999_7108054 | -0.81 | 5.2e-02 | Click! |
Activity profile of RREB1 motif
Sorted Z-values of RREB1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_96588279 | 0.31 |
ENST00000552142.1
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr17_-_58499766 | 0.21 |
ENST00000588898.1
|
USP32
|
ubiquitin specific peptidase 32 |
| chr14_+_91709279 | 0.20 |
ENST00000554096.1
|
CTD-2547L24.3
|
HCG1816139; Uncharacterized protein |
| chr3_+_141205852 | 0.18 |
ENST00000286364.3
ENST00000452898.1 |
RASA2
|
RAS p21 protein activator 2 |
| chr11_-_129872672 | 0.17 |
ENST00000531431.1
ENST00000527581.1 |
PRDM10
|
PR domain containing 10 |
| chr20_+_62697564 | 0.17 |
ENST00000458442.1
|
TCEA2
|
transcription elongation factor A (SII), 2 |
| chr11_-_96239990 | 0.16 |
ENST00000511243.2
|
JRKL-AS1
|
JRKL antisense RNA 1 |
| chr2_+_242312264 | 0.16 |
ENST00000445489.1
|
FARP2
|
FERM, RhoGEF and pleckstrin domain protein 2 |
| chr17_-_39216344 | 0.15 |
ENST00000391418.2
|
KRTAP2-3
|
keratin associated protein 2-3 |
| chr3_+_192958914 | 0.15 |
ENST00000264735.2
ENST00000602513.1 |
HRASLS
|
HRAS-like suppressor |
| chr17_+_1733251 | 0.14 |
ENST00000570451.1
|
RPA1
|
replication protein A1, 70kDa |
| chr22_+_41968007 | 0.14 |
ENST00000460790.1
|
CSDC2
|
cold shock domain containing C2, RNA binding |
| chr11_+_111896090 | 0.14 |
ENST00000393051.1
|
DLAT
|
dihydrolipoamide S-acetyltransferase |
| chrX_-_48693955 | 0.13 |
ENST00000218230.5
|
PCSK1N
|
proprotein convertase subtilisin/kexin type 1 inhibitor |
| chr3_+_41241596 | 0.13 |
ENST00000450969.1
|
CTNNB1
|
catenin (cadherin-associated protein), beta 1, 88kDa |
| chr1_+_16083098 | 0.13 |
ENST00000496928.2
ENST00000508310.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chrX_-_119249819 | 0.12 |
ENST00000217999.2
|
RHOXF1
|
Rhox homeobox family, member 1 |
| chr13_-_74708372 | 0.12 |
ENST00000377666.4
|
KLF12
|
Kruppel-like factor 12 |
| chr4_-_87374330 | 0.11 |
ENST00000511328.1
ENST00000503911.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr18_-_21242833 | 0.11 |
ENST00000586087.1
ENST00000592179.1 |
ANKRD29
|
ankyrin repeat domain 29 |
| chr18_-_21242729 | 0.11 |
ENST00000585908.2
|
ANKRD29
|
ankyrin repeat domain 29 |
| chr16_-_3285144 | 0.11 |
ENST00000431561.3
ENST00000396870.4 |
ZNF200
|
zinc finger protein 200 |
| chr19_-_51456321 | 0.10 |
ENST00000391809.2
|
KLK5
|
kallikrein-related peptidase 5 |
| chr9_+_132427883 | 0.10 |
ENST00000372469.4
|
PRRX2
|
paired related homeobox 2 |
| chr19_-_51456198 | 0.10 |
ENST00000594846.1
|
KLK5
|
kallikrein-related peptidase 5 |
| chr19_-_51456344 | 0.10 |
ENST00000336334.3
ENST00000593428.1 |
KLK5
|
kallikrein-related peptidase 5 |
| chr8_+_144371773 | 0.10 |
ENST00000523891.1
|
ZNF696
|
zinc finger protein 696 |
| chr3_-_52869205 | 0.10 |
ENST00000446157.2
|
MUSTN1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr12_-_48213735 | 0.10 |
ENST00000417902.1
ENST00000417107.1 |
HDAC7
|
histone deacetylase 7 |
| chr11_+_111896320 | 0.09 |
ENST00000531306.1
ENST00000537636.1 |
DLAT
|
dihydrolipoamide S-acetyltransferase |
| chrX_+_17755563 | 0.09 |
ENST00000380045.3
ENST00000380041.3 ENST00000380043.3 ENST00000398080.1 |
SCML1
|
sex comb on midleg-like 1 (Drosophila) |
| chrX_-_100604184 | 0.09 |
ENST00000372902.3
|
TIMM8A
|
translocase of inner mitochondrial membrane 8 homolog A (yeast) |
| chr13_-_52026730 | 0.09 |
ENST00000420668.2
|
INTS6
|
integrator complex subunit 6 |
| chr16_-_15736345 | 0.09 |
ENST00000549219.1
|
KIAA0430
|
KIAA0430 |
| chr12_-_62653903 | 0.09 |
ENST00000552075.1
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
| chr1_-_228604454 | 0.09 |
ENST00000456946.2
|
TRIM17
|
tripartite motif containing 17 |
| chr7_-_19157248 | 0.08 |
ENST00000242261.5
|
TWIST1
|
twist family bHLH transcription factor 1 |
| chr3_+_154797636 | 0.08 |
ENST00000481828.1
|
MME
|
membrane metallo-endopeptidase |
| chr13_-_77601282 | 0.08 |
ENST00000355619.5
|
FBXL3
|
F-box and leucine-rich repeat protein 3 |
| chr17_-_39728303 | 0.08 |
ENST00000588431.1
ENST00000246662.4 |
KRT9
|
keratin 9 |
| chr2_-_179343226 | 0.08 |
ENST00000434643.2
|
FKBP7
|
FK506 binding protein 7 |
| chr4_-_56412713 | 0.08 |
ENST00000435527.2
|
CLOCK
|
clock circadian regulator |
| chr2_-_67442409 | 0.08 |
ENST00000414404.1
ENST00000433133.1 |
AC078941.1
|
AC078941.1 |
| chr19_+_42381173 | 0.08 |
ENST00000221972.3
|
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr16_-_85146040 | 0.08 |
ENST00000539556.1
|
FAM92B
|
family with sequence similarity 92, member B |
| chr3_-_42003479 | 0.08 |
ENST00000420927.1
|
ULK4
|
unc-51 like kinase 4 |
| chr11_-_61196858 | 0.08 |
ENST00000413184.2
|
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa |
| chr2_-_37551846 | 0.08 |
ENST00000443187.1
|
PRKD3
|
protein kinase D3 |
| chr20_-_1165117 | 0.08 |
ENST00000381894.3
|
TMEM74B
|
transmembrane protein 74B |
| chr8_+_90914757 | 0.08 |
ENST00000451899.2
|
OSGIN2
|
oxidative stress induced growth inhibitor family member 2 |
| chr8_+_26149274 | 0.08 |
ENST00000522535.1
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr21_-_34915123 | 0.07 |
ENST00000438059.1
|
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr10_+_115438920 | 0.07 |
ENST00000429617.1
ENST00000369331.4 |
CASP7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr12_-_131323719 | 0.07 |
ENST00000392373.2
|
STX2
|
syntaxin 2 |
| chr17_+_30348024 | 0.07 |
ENST00000327564.7
ENST00000584368.1 ENST00000394713.3 ENST00000341671.7 |
LRRC37B
|
leucine rich repeat containing 37B |
| chr22_-_46450024 | 0.07 |
ENST00000396008.2
ENST00000333761.1 |
C22orf26
|
chromosome 22 open reading frame 26 |
| chr16_+_28889801 | 0.07 |
ENST00000395503.4
|
ATP2A1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr5_+_78908388 | 0.07 |
ENST00000296783.3
|
PAPD4
|
PAP associated domain containing 4 |
| chr4_+_75230853 | 0.07 |
ENST00000244869.2
|
EREG
|
epiregulin |
| chrX_+_128674213 | 0.07 |
ENST00000371113.4
ENST00000357121.5 |
OCRL
|
oculocerebrorenal syndrome of Lowe |
| chr12_-_8088773 | 0.07 |
ENST00000544291.1
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr17_-_62658186 | 0.07 |
ENST00000262435.9
|
SMURF2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr19_-_49140609 | 0.07 |
ENST00000601104.1
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr12_-_53297432 | 0.07 |
ENST00000546900.1
|
KRT8
|
keratin 8 |
| chr12_-_44152551 | 0.07 |
ENST00000416848.2
ENST00000550784.1 ENST00000547156.1 ENST00000549868.1 ENST00000553166.1 ENST00000551923.1 ENST00000431332.3 ENST00000344862.5 |
PUS7L
|
pseudouridylate synthase 7 homolog (S. cerevisiae)-like |
| chrX_-_131352152 | 0.07 |
ENST00000342983.2
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr6_-_13486369 | 0.07 |
ENST00000558378.1
|
AL583828.1
|
AL583828.1 |
| chr9_+_93921855 | 0.06 |
ENST00000423719.2
ENST00000609752.1 |
LINC00484
|
long intergenic non-protein coding RNA 484 |
| chr19_-_47975143 | 0.06 |
ENST00000597014.1
|
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chr10_+_127661942 | 0.06 |
ENST00000417114.1
ENST00000445510.1 ENST00000368691.1 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr12_-_131323754 | 0.06 |
ENST00000261653.6
|
STX2
|
syntaxin 2 |
| chr19_-_51071302 | 0.06 |
ENST00000389201.3
ENST00000600381.1 |
LRRC4B
|
leucine rich repeat containing 4B |
| chr17_+_41924536 | 0.06 |
ENST00000317310.4
|
CD300LG
|
CD300 molecule-like family member g |
| chr1_+_46668994 | 0.06 |
ENST00000371980.3
|
LURAP1
|
leucine rich adaptor protein 1 |
| chr7_+_114562172 | 0.06 |
ENST00000393486.1
ENST00000257724.3 |
MDFIC
|
MyoD family inhibitor domain containing |
| chr5_+_92228 | 0.06 |
ENST00000512035.1
|
CTD-2231H16.1
|
CTD-2231H16.1 |
| chr16_-_1275257 | 0.06 |
ENST00000234798.4
|
TPSG1
|
tryptase gamma 1 |
| chr1_-_201346761 | 0.06 |
ENST00000455702.1
ENST00000422165.1 ENST00000367318.5 ENST00000367320.2 ENST00000438742.1 ENST00000412633.1 ENST00000458432.2 ENST00000421663.2 ENST00000367322.1 ENST00000509001.1 |
TNNT2
|
troponin T type 2 (cardiac) |
| chr10_-_112064665 | 0.06 |
ENST00000369603.5
|
SMNDC1
|
survival motor neuron domain containing 1 |
| chr14_+_57735614 | 0.06 |
ENST00000261558.3
|
AP5M1
|
adaptor-related protein complex 5, mu 1 subunit |
| chr17_-_46703826 | 0.06 |
ENST00000550387.1
ENST00000311177.5 |
HOXB9
|
homeobox B9 |
| chr2_+_48757278 | 0.06 |
ENST00000404752.1
ENST00000406226.1 |
STON1
|
stonin 1 |
| chr7_+_95401877 | 0.06 |
ENST00000524053.1
ENST00000324972.6 ENST00000537881.1 ENST00000437599.1 ENST00000359388.4 ENST00000413338.1 |
DYNC1I1
|
dynein, cytoplasmic 1, intermediate chain 1 |
| chr11_-_299519 | 0.06 |
ENST00000382614.2
|
IFITM5
|
interferon induced transmembrane protein 5 |
| chr2_+_102686820 | 0.05 |
ENST00000409929.1
ENST00000424272.1 |
IL1R1
|
interleukin 1 receptor, type I |
| chr9_-_140335789 | 0.05 |
ENST00000344119.2
ENST00000371506.2 |
ENTPD8
|
ectonucleoside triphosphate diphosphohydrolase 8 |
| chr13_-_37494391 | 0.05 |
ENST00000379826.4
|
SMAD9
|
SMAD family member 9 |
| chr9_+_136215087 | 0.05 |
ENST00000426651.1
|
RPL7A
|
ribosomal protein L7a |
| chr8_+_90914859 | 0.05 |
ENST00000520659.1
|
OSGIN2
|
oxidative stress induced growth inhibitor family member 2 |
| chr12_-_95044309 | 0.05 |
ENST00000261226.4
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr16_+_28303804 | 0.05 |
ENST00000341901.4
|
SBK1
|
SH3 domain binding kinase 1 |
| chr17_-_7082861 | 0.05 |
ENST00000269299.3
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr7_+_97840739 | 0.05 |
ENST00000609256.1
|
BHLHA15
|
basic helix-loop-helix family, member a15 |
| chr6_-_86353510 | 0.05 |
ENST00000444272.1
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr16_+_30969055 | 0.05 |
ENST00000452917.1
|
SETD1A
|
SET domain containing 1A |
| chr17_-_38978847 | 0.05 |
ENST00000269576.5
|
KRT10
|
keratin 10 |
| chr2_+_46926326 | 0.05 |
ENST00000394861.2
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr16_+_28889703 | 0.05 |
ENST00000357084.3
|
ATP2A1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr17_+_7788104 | 0.05 |
ENST00000380358.4
|
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chrX_-_148571884 | 0.05 |
ENST00000537071.1
|
IDS
|
iduronate 2-sulfatase |
| chr8_-_66753682 | 0.05 |
ENST00000396642.3
|
PDE7A
|
phosphodiesterase 7A |
| chrX_+_17755696 | 0.05 |
ENST00000419185.1
|
SCML1
|
sex comb on midleg-like 1 (Drosophila) |
| chr5_+_78908233 | 0.05 |
ENST00000453514.1
ENST00000423041.2 ENST00000504233.1 ENST00000428308.2 |
PAPD4
|
PAP associated domain containing 4 |
| chr14_-_71067360 | 0.05 |
ENST00000554963.1
ENST00000430055.2 ENST00000440435.2 ENST00000256379.5 |
MED6
|
mediator complex subunit 6 |
| chr1_+_110091189 | 0.05 |
ENST00000369851.4
|
GNAI3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 |
| chr16_-_49890016 | 0.05 |
ENST00000563137.2
|
ZNF423
|
zinc finger protein 423 |
| chr8_+_95653840 | 0.05 |
ENST00000520385.1
|
ESRP1
|
epithelial splicing regulatory protein 1 |
| chr16_+_14396121 | 0.05 |
ENST00000570945.1
|
RP11-65J21.3
|
RP11-65J21.3 |
| chr2_+_223289208 | 0.05 |
ENST00000321276.7
|
SGPP2
|
sphingosine-1-phosphate phosphatase 2 |
| chr1_-_153538292 | 0.05 |
ENST00000497140.1
ENST00000368708.3 |
S100A2
|
S100 calcium binding protein A2 |
| chr10_-_71930222 | 0.05 |
ENST00000458634.2
ENST00000373239.2 ENST00000373242.2 ENST00000373241.4 |
SAR1A
|
SAR1 homolog A (S. cerevisiae) |
| chr17_-_1733114 | 0.05 |
ENST00000305513.7
|
SMYD4
|
SET and MYND domain containing 4 |
| chr9_-_116163400 | 0.05 |
ENST00000277315.5
ENST00000448137.1 ENST00000409155.3 |
ALAD
|
aminolevulinate dehydratase |
| chr5_+_151151471 | 0.05 |
ENST00000394123.3
ENST00000543466.1 |
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr19_-_39226045 | 0.04 |
ENST00000597987.1
ENST00000595177.1 |
CAPN12
|
calpain 12 |
| chr13_+_24734880 | 0.04 |
ENST00000382095.4
|
SPATA13
|
spermatogenesis associated 13 |
| chr8_+_38261880 | 0.04 |
ENST00000527175.1
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr12_-_6716569 | 0.04 |
ENST00000544040.1
ENST00000545942.1 |
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr8_-_75233563 | 0.04 |
ENST00000342232.4
|
JPH1
|
junctophilin 1 |
| chr17_-_6459802 | 0.04 |
ENST00000262483.8
|
PITPNM3
|
PITPNM family member 3 |
| chr12_-_48226897 | 0.04 |
ENST00000434070.1
|
HDAC7
|
histone deacetylase 7 |
| chr12_-_123756781 | 0.04 |
ENST00000544658.1
|
CDK2AP1
|
cyclin-dependent kinase 2 associated protein 1 |
| chr6_+_37321823 | 0.04 |
ENST00000487950.1
ENST00000469731.1 |
RNF8
|
ring finger protein 8, E3 ubiquitin protein ligase |
| chr17_+_18761417 | 0.04 |
ENST00000419284.2
ENST00000268835.2 ENST00000412418.1 ENST00000575228.1 ENST00000575102.1 ENST00000536323.1 |
PRPSAP2
|
phosphoribosyl pyrophosphate synthetase-associated protein 2 |
| chr22_-_36357671 | 0.04 |
ENST00000408983.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr12_-_123756687 | 0.04 |
ENST00000261692.2
|
CDK2AP1
|
cyclin-dependent kinase 2 associated protein 1 |
| chr12_+_56415100 | 0.04 |
ENST00000547791.1
|
IKZF4
|
IKAROS family zinc finger 4 (Eos) |
| chr17_+_35849937 | 0.04 |
ENST00000394389.4
|
DUSP14
|
dual specificity phosphatase 14 |
| chr14_+_91709103 | 0.04 |
ENST00000553725.1
|
CTD-2547L24.3
|
HCG1816139; Uncharacterized protein |
| chr2_-_74645669 | 0.04 |
ENST00000518401.1
|
C2orf81
|
chromosome 2 open reading frame 81 |
| chr1_-_113392399 | 0.04 |
ENST00000449572.2
ENST00000433505.1 |
RP11-426L16.8
|
RP11-426L16.8 |
| chr18_+_9334786 | 0.04 |
ENST00000581641.1
|
TWSG1
|
twisted gastrulation BMP signaling modulator 1 |
| chr6_+_37321748 | 0.04 |
ENST00000373479.4
ENST00000394443.4 |
RNF8
|
ring finger protein 8, E3 ubiquitin protein ligase |
| chr7_-_24797032 | 0.04 |
ENST00000409970.1
ENST00000409775.3 |
DFNA5
|
deafness, autosomal dominant 5 |
| chr10_-_95360983 | 0.04 |
ENST00000371464.3
|
RBP4
|
retinol binding protein 4, plasma |
| chr19_-_44084586 | 0.04 |
ENST00000599693.1
ENST00000598165.1 |
XRCC1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr14_+_74353320 | 0.04 |
ENST00000540593.1
ENST00000555730.1 |
ZNF410
|
zinc finger protein 410 |
| chr5_+_125758865 | 0.04 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr4_+_56719782 | 0.04 |
ENST00000381295.2
ENST00000346134.7 ENST00000349598.6 |
EXOC1
|
exocyst complex component 1 |
| chr5_-_133512654 | 0.04 |
ENST00000522552.1
|
SKP1
|
S-phase kinase-associated protein 1 |
| chr11_-_74109422 | 0.04 |
ENST00000298198.4
|
PGM2L1
|
phosphoglucomutase 2-like 1 |
| chr12_-_56101647 | 0.04 |
ENST00000347027.6
ENST00000257879.6 ENST00000257880.7 ENST00000394230.2 ENST00000394229.2 |
ITGA7
|
integrin, alpha 7 |
| chrX_-_151307020 | 0.04 |
ENST00000370323.4
ENST00000244096.3 ENST00000427322.2 |
MAGEA10
|
melanoma antigen family A, 10 |
| chr11_-_35547277 | 0.04 |
ENST00000527605.1
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr16_+_21169976 | 0.04 |
ENST00000572258.1
ENST00000261388.3 ENST00000451578.2 ENST00000572599.1 ENST00000577162.1 |
TMEM159
|
transmembrane protein 159 |
| chr6_-_86352982 | 0.04 |
ENST00000369622.3
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr20_+_33462745 | 0.04 |
ENST00000488172.1
|
ACSS2
|
acyl-CoA synthetase short-chain family member 2 |
| chr17_+_55334364 | 0.04 |
ENST00000322684.3
ENST00000579590.1 |
MSI2
|
musashi RNA-binding protein 2 |
| chr7_+_1022811 | 0.04 |
ENST00000308919.7
|
CYP2W1
|
cytochrome P450, family 2, subfamily W, polypeptide 1 |
| chr8_-_145578505 | 0.04 |
ENST00000526263.1
ENST00000398633.3 |
TMEM249
|
transmembrane protein 249 |
| chr5_+_151151504 | 0.04 |
ENST00000356245.3
ENST00000507878.2 |
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr13_-_108867846 | 0.04 |
ENST00000442234.1
|
LIG4
|
ligase IV, DNA, ATP-dependent |
| chr17_-_66597530 | 0.04 |
ENST00000592554.1
|
FAM20A
|
family with sequence similarity 20, member A |
| chr11_-_85780086 | 0.04 |
ENST00000532317.1
ENST00000528256.1 ENST00000526033.1 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr17_+_38296576 | 0.04 |
ENST00000264645.7
|
CASC3
|
cancer susceptibility candidate 3 |
| chr11_+_34073195 | 0.04 |
ENST00000341394.4
|
CAPRIN1
|
cell cycle associated protein 1 |
| chr4_+_74735102 | 0.04 |
ENST00000395761.3
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
| chr11_+_128562372 | 0.04 |
ENST00000344954.6
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr19_-_30206128 | 0.04 |
ENST00000392276.1
ENST00000592153.1 ENST00000323670.9 |
C19orf12
|
chromosome 19 open reading frame 12 |
| chr5_-_133512683 | 0.04 |
ENST00000353411.6
|
SKP1
|
S-phase kinase-associated protein 1 |
| chr14_+_74353574 | 0.04 |
ENST00000442160.3
ENST00000555044.1 |
ZNF410
|
zinc finger protein 410 |
| chr12_-_6716534 | 0.04 |
ENST00000544484.1
ENST00000309577.6 ENST00000357008.2 |
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr5_-_124080203 | 0.04 |
ENST00000504926.1
|
ZNF608
|
zinc finger protein 608 |
| chr17_-_66453562 | 0.04 |
ENST00000262139.5
ENST00000546360.1 |
WIPI1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr7_+_152456829 | 0.03 |
ENST00000377776.3
ENST00000256001.8 ENST00000397282.2 |
ACTR3B
|
ARP3 actin-related protein 3 homolog B (yeast) |
| chr11_-_8739566 | 0.03 |
ENST00000533020.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr7_-_92465868 | 0.03 |
ENST00000424848.2
|
CDK6
|
cyclin-dependent kinase 6 |
| chr11_-_61197406 | 0.03 |
ENST00000541963.1
ENST00000477890.2 |
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa |
| chr2_-_32236002 | 0.03 |
ENST00000404530.1
|
MEMO1
|
mediator of cell motility 1 |
| chr18_-_21242774 | 0.03 |
ENST00000322980.9
|
ANKRD29
|
ankyrin repeat domain 29 |
| chr11_+_17756279 | 0.03 |
ENST00000265969.6
|
KCNC1
|
potassium voltage-gated channel, Shaw-related subfamily, member 1 |
| chr16_+_1728257 | 0.03 |
ENST00000248098.3
ENST00000562684.1 ENST00000561516.1 ENST00000382711.5 ENST00000566742.1 |
HN1L
|
hematological and neurological expressed 1-like |
| chr13_-_37494365 | 0.03 |
ENST00000350148.5
|
SMAD9
|
SMAD family member 9 |
| chr12_-_57940904 | 0.03 |
ENST00000550954.1
ENST00000434715.3 ENST00000546670.1 ENST00000543672.1 |
DCTN2
|
dynactin 2 (p50) |
| chr11_+_125034586 | 0.03 |
ENST00000298282.9
|
PKNOX2
|
PBX/knotted 1 homeobox 2 |
| chr1_+_220267429 | 0.03 |
ENST00000366922.1
ENST00000302637.5 |
IARS2
|
isoleucyl-tRNA synthetase 2, mitochondrial |
| chr6_+_112375462 | 0.03 |
ENST00000361714.1
|
WISP3
|
WNT1 inducible signaling pathway protein 3 |
| chr9_-_13279589 | 0.03 |
ENST00000319217.7
|
MPDZ
|
multiple PDZ domain protein |
| chr17_+_1733276 | 0.03 |
ENST00000254719.5
|
RPA1
|
replication protein A1, 70kDa |
| chr5_+_125758813 | 0.03 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr12_-_12715266 | 0.03 |
ENST00000228862.2
|
DUSP16
|
dual specificity phosphatase 16 |
| chr12_+_44152740 | 0.03 |
ENST00000440781.2
ENST00000431837.1 ENST00000550616.1 ENST00000448290.2 ENST00000551736.1 |
IRAK4
|
interleukin-1 receptor-associated kinase 4 |
| chr11_+_1093318 | 0.03 |
ENST00000333592.6
|
MUC2
|
mucin 2, oligomeric mucus/gel-forming |
| chr2_+_192542850 | 0.03 |
ENST00000410026.2
|
NABP1
|
nucleic acid binding protein 1 |
| chr9_-_13279563 | 0.03 |
ENST00000541718.1
|
MPDZ
|
multiple PDZ domain protein |
| chr13_+_24734844 | 0.03 |
ENST00000382108.3
|
SPATA13
|
spermatogenesis associated 13 |
| chr2_-_54087066 | 0.03 |
ENST00000394705.2
ENST00000352846.3 ENST00000406625.2 |
GPR75
GPR75-ASB3
ASB3
|
G protein-coupled receptor 75 GPR75-ASB3 readthrough Ankyrin repeat and SOCS box protein 3 |
| chr11_+_61197572 | 0.03 |
ENST00000542074.1
ENST00000534878.1 ENST00000537782.1 ENST00000543265.1 |
SDHAF2
|
succinate dehydrogenase complex assembly factor 2 |
| chr16_-_4588822 | 0.03 |
ENST00000564828.1
|
CDIP1
|
cell death-inducing p53 target 1 |
| chrX_-_153979315 | 0.03 |
ENST00000369575.3
ENST00000369568.4 ENST00000424127.2 |
GAB3
|
GRB2-associated binding protein 3 |
| chr2_+_192542879 | 0.03 |
ENST00000409510.1
|
NABP1
|
nucleic acid binding protein 1 |
| chr11_+_61197508 | 0.03 |
ENST00000541135.1
ENST00000301761.2 |
RP11-286N22.8
SDHAF2
|
Uncharacterized protein succinate dehydrogenase complex assembly factor 2 |
| chr7_-_752074 | 0.03 |
ENST00000360274.4
|
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr9_+_96026230 | 0.03 |
ENST00000448251.1
|
WNK2
|
WNK lysine deficient protein kinase 2 |
| chr20_+_816695 | 0.03 |
ENST00000246100.3
|
FAM110A
|
family with sequence similarity 110, member A |
| chr17_-_73840415 | 0.03 |
ENST00000592386.1
ENST00000412096.2 ENST00000586147.1 |
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr16_+_67143828 | 0.03 |
ENST00000563853.2
ENST00000569914.1 ENST00000569600.1 |
C16orf70
|
chromosome 16 open reading frame 70 |
| chr12_+_62654155 | 0.03 |
ENST00000312635.6
ENST00000393654.3 ENST00000549237.1 |
USP15
|
ubiquitin specific peptidase 15 |
| chr11_-_85779971 | 0.03 |
ENST00000393346.3
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr2_-_61765315 | 0.03 |
ENST00000406957.1
ENST00000401558.2 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
Network of associatons between targets according to the STRING database.
First level regulatory network of RREB1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.0 | 0.2 | GO:1904499 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.0 | 0.1 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.0 | 0.1 | GO:2000276 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.1 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.0 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.3 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.0 | GO:1904875 | regulation of DNA ligase activity(GO:1904875) |
| 0.0 | 0.1 | GO:0071484 | cellular response to light intensity(GO:0071484) |
| 0.0 | 0.0 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.2 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.0 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.0 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.0 | 0.0 | GO:0071030 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.0 | 0.1 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.0 | GO:0002432 | granuloma formation(GO:0002432) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.3 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.2 | GO:0070369 | Scrib-APC-beta-catenin complex(GO:0034750) beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.0 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.0 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.0 | GO:0070701 | mucus layer(GO:0070701) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.1 | 0.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0004641 | phosphoribosylamine-glycine ligase activity(GO:0004637) phosphoribosylformylglycinamidine cyclo-ligase activity(GO:0004641) phosphoribosylglycinamide formyltransferase activity(GO:0004644) |
| 0.0 | 0.0 | GO:0004423 | iduronate-2-sulfatase activity(GO:0004423) |
| 0.0 | 0.0 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.0 | 0.0 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.0 | 0.0 | GO:0098770 | FBXO family protein binding(GO:0098770) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |