Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
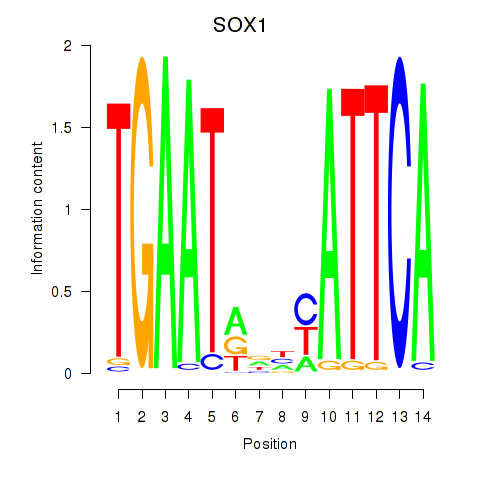
Results for SOX1
Z-value: 0.54
Transcription factors associated with SOX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX1
|
ENSG00000182968.3 | SRY-box transcription factor 1 |
Activity profile of SOX1 motif
Sorted Z-values of SOX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_76955563 | 0.40 |
ENST00000441833.2
|
GSAP
|
gamma-secretase activating protein |
| chr19_-_50990785 | 0.33 |
ENST00000595005.1
|
CTD-2545M3.8
|
CTD-2545M3.8 |
| chr17_+_74729060 | 0.24 |
ENST00000587459.1
|
RP11-318A15.7
|
Uncharacterized protein |
| chr12_+_55248289 | 0.20 |
ENST00000308796.6
|
MUCL1
|
mucin-like 1 |
| chr17_+_80816395 | 0.19 |
ENST00000576160.2
ENST00000571712.1 |
TBCD
|
tubulin folding cofactor D |
| chr6_+_148593425 | 0.19 |
ENST00000367469.1
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr3_-_55539287 | 0.17 |
ENST00000472238.1
|
RP11-875H7.5
|
RP11-875H7.5 |
| chr6_+_26183958 | 0.17 |
ENST00000356530.3
|
HIST1H2BE
|
histone cluster 1, H2be |
| chr3_-_126327398 | 0.16 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr19_-_44388116 | 0.15 |
ENST00000587539.1
|
ZNF404
|
zinc finger protein 404 |
| chr2_+_171034646 | 0.14 |
ENST00000409044.3
ENST00000408978.4 |
MYO3B
|
myosin IIIB |
| chr14_+_35514323 | 0.14 |
ENST00000555211.1
|
FAM177A1
|
family with sequence similarity 177, member A1 |
| chr1_+_32674675 | 0.14 |
ENST00000409358.1
|
DCDC2B
|
doublecortin domain containing 2B |
| chr11_+_125496124 | 0.13 |
ENST00000533778.2
ENST00000534070.1 |
CHEK1
|
checkpoint kinase 1 |
| chr21_-_23058648 | 0.13 |
ENST00000416182.1
|
AF241725.6
|
AF241725.6 |
| chr10_-_92681033 | 0.13 |
ENST00000371697.3
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr1_+_152957707 | 0.13 |
ENST00000368762.1
|
SPRR1A
|
small proline-rich protein 1A |
| chr8_-_86290333 | 0.13 |
ENST00000521846.1
ENST00000523022.1 ENST00000524324.1 ENST00000519991.1 ENST00000520663.1 ENST00000517590.1 ENST00000522579.1 ENST00000522814.1 ENST00000522662.1 ENST00000523858.1 ENST00000519129.1 |
CA1
|
carbonic anhydrase I |
| chr6_-_15548591 | 0.12 |
ENST00000509674.1
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr12_+_9980069 | 0.12 |
ENST00000354855.3
ENST00000324214.4 ENST00000279544.3 |
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1 |
| chr15_+_42565393 | 0.12 |
ENST00000561871.1
|
GANC
|
glucosidase, alpha; neutral C |
| chr8_-_134501873 | 0.11 |
ENST00000523634.1
|
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr2_-_96926313 | 0.11 |
ENST00000435268.1
|
TMEM127
|
transmembrane protein 127 |
| chr2_-_14541060 | 0.11 |
ENST00000418420.1
ENST00000417751.1 |
LINC00276
|
long intergenic non-protein coding RNA 276 |
| chr4_+_70861647 | 0.10 |
ENST00000246895.4
ENST00000381060.2 |
STATH
|
statherin |
| chr11_-_18062872 | 0.09 |
ENST00000250018.2
|
TPH1
|
tryptophan hydroxylase 1 |
| chr14_+_38033252 | 0.09 |
ENST00000554829.1
|
RP11-356O9.1
|
RP11-356O9.1 |
| chr19_+_21324827 | 0.09 |
ENST00000600692.1
ENST00000599296.1 ENST00000594425.1 ENST00000311048.7 |
ZNF431
|
zinc finger protein 431 |
| chr11_+_29181503 | 0.09 |
ENST00000530960.1
|
RP11-466I1.1
|
RP11-466I1.1 |
| chr3_+_132316081 | 0.09 |
ENST00000249887.2
|
ACKR4
|
atypical chemokine receptor 4 |
| chr9_-_85882145 | 0.09 |
ENST00000328788.1
|
FRMD3
|
FERM domain containing 3 |
| chr5_+_158654712 | 0.08 |
ENST00000520323.1
|
CTB-11I22.2
|
CTB-11I22.2 |
| chr17_-_15519008 | 0.08 |
ENST00000261644.8
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr8_+_145133493 | 0.08 |
ENST00000316052.5
ENST00000525936.1 |
EXOSC4
|
exosome component 4 |
| chr11_-_26593677 | 0.07 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr1_+_24646263 | 0.07 |
ENST00000524724.1
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr7_+_36450169 | 0.07 |
ENST00000428612.1
|
ANLN
|
anillin, actin binding protein |
| chr20_+_49575342 | 0.07 |
ENST00000244051.1
|
MOCS3
|
molybdenum cofactor synthesis 3 |
| chr12_-_10607084 | 0.06 |
ENST00000408006.3
ENST00000544822.1 ENST00000536188.1 |
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr12_-_11150474 | 0.06 |
ENST00000538986.1
|
TAS2R20
|
taste receptor, type 2, member 20 |
| chr12_+_93096619 | 0.06 |
ENST00000397833.3
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr10_+_23384435 | 0.06 |
ENST00000376510.3
|
MSRB2
|
methionine sulfoxide reductase B2 |
| chr6_-_138866823 | 0.06 |
ENST00000342260.5
|
NHSL1
|
NHS-like 1 |
| chr19_+_49588690 | 0.06 |
ENST00000221448.5
|
SNRNP70
|
small nuclear ribonucleoprotein 70kDa (U1) |
| chr3_-_58200398 | 0.06 |
ENST00000318316.3
ENST00000460422.1 ENST00000483681.1 |
DNASE1L3
|
deoxyribonuclease I-like 3 |
| chr19_-_10491130 | 0.06 |
ENST00000530829.1
ENST00000529370.1 |
TYK2
|
tyrosine kinase 2 |
| chr1_-_237167718 | 0.06 |
ENST00000464121.2
|
MT1HL1
|
metallothionein 1H-like 1 |
| chr10_+_15085895 | 0.06 |
ENST00000378228.3
|
OLAH
|
oleoyl-ACP hydrolase |
| chr3_-_107596910 | 0.06 |
ENST00000464359.2
ENST00000464823.1 ENST00000466155.1 ENST00000473528.2 ENST00000608306.1 ENST00000488852.1 ENST00000608137.1 ENST00000608307.1 ENST00000609429.1 ENST00000601385.1 ENST00000475362.1 ENST00000600240.1 ENST00000600749.1 |
LINC00635
|
long intergenic non-protein coding RNA 635 |
| chr3_-_44519131 | 0.06 |
ENST00000425708.2
ENST00000396077.2 |
ZNF445
|
zinc finger protein 445 |
| chr2_+_171036635 | 0.05 |
ENST00000484338.2
ENST00000334231.6 |
MYO3B
|
myosin IIIB |
| chr1_-_52499443 | 0.05 |
ENST00000371614.1
|
KTI12
|
KTI12 homolog, chromatin associated (S. cerevisiae) |
| chr2_+_11864458 | 0.05 |
ENST00000396098.1
ENST00000396099.1 ENST00000425416.2 |
LPIN1
|
lipin 1 |
| chr2_+_87808725 | 0.05 |
ENST00000413202.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr3_-_185655795 | 0.04 |
ENST00000342294.4
ENST00000382191.4 ENST00000453386.2 |
TRA2B
|
transformer 2 beta homolog (Drosophila) |
| chr11_-_57177586 | 0.04 |
ENST00000529411.1
|
RP11-872D17.8
|
Uncharacterized protein |
| chr12_-_10605929 | 0.04 |
ENST00000347831.5
ENST00000359151.3 |
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr5_-_54988559 | 0.04 |
ENST00000502247.1
|
SLC38A9
|
solute carrier family 38, member 9 |
| chr8_-_134501937 | 0.04 |
ENST00000519924.1
|
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr19_+_17830051 | 0.04 |
ENST00000594625.1
ENST00000324096.4 ENST00000600186.1 ENST00000597735.1 |
MAP1S
|
microtubule-associated protein 1S |
| chr9_-_123638633 | 0.04 |
ENST00000456291.1
|
PHF19
|
PHD finger protein 19 |
| chr2_+_234296792 | 0.04 |
ENST00000409813.3
|
DGKD
|
diacylglycerol kinase, delta 130kDa |
| chr6_+_27833034 | 0.03 |
ENST00000357320.2
|
HIST1H2AL
|
histone cluster 1, H2al |
| chr11_-_26593649 | 0.03 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chr22_-_17302589 | 0.03 |
ENST00000331428.5
|
XKR3
|
XK, Kell blood group complex subunit-related family, member 3 |
| chrX_-_57021943 | 0.03 |
ENST00000374919.3
|
SPIN3
|
spindlin family, member 3 |
| chr6_+_130742231 | 0.03 |
ENST00000545622.1
|
TMEM200A
|
transmembrane protein 200A |
| chr1_+_202789394 | 0.03 |
ENST00000330493.5
|
RP11-480I12.4
|
Putative inactive alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase-like protein LOC641515 |
| chr6_-_130543958 | 0.03 |
ENST00000437477.2
ENST00000439090.2 |
SAMD3
|
sterile alpha motif domain containing 3 |
| chr14_+_62462541 | 0.03 |
ENST00000430451.2
|
SYT16
|
synaptotagmin XVI |
| chr16_-_11876408 | 0.03 |
ENST00000396516.2
|
ZC3H7A
|
zinc finger CCCH-type containing 7A |
| chr15_+_57540230 | 0.03 |
ENST00000559703.1
|
TCF12
|
transcription factor 12 |
| chr17_-_38821373 | 0.03 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr19_+_49588677 | 0.02 |
ENST00000598984.1
ENST00000598441.1 |
SNRNP70
|
small nuclear ribonucleoprotein 70kDa (U1) |
| chr17_-_39646116 | 0.02 |
ENST00000328119.6
|
KRT36
|
keratin 36 |
| chr1_+_174769006 | 0.02 |
ENST00000489615.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr13_-_22178284 | 0.02 |
ENST00000468222.2
ENST00000382374.4 |
MICU2
|
mitochondrial calcium uptake 2 |
| chr11_+_55029628 | 0.02 |
ENST00000417545.2
|
TRIM48
|
tripartite motif containing 48 |
| chr17_+_75316336 | 0.02 |
ENST00000591934.1
|
SEPT9
|
septin 9 |
| chr11_-_78052923 | 0.02 |
ENST00000340149.2
|
GAB2
|
GRB2-associated binding protein 2 |
| chr10_-_73976025 | 0.02 |
ENST00000342444.4
ENST00000533958.1 ENST00000527593.1 ENST00000394915.3 ENST00000530461.1 ENST00000317168.6 ENST00000524829.1 |
ASCC1
|
activating signal cointegrator 1 complex subunit 1 |
| chr5_+_175740083 | 0.02 |
ENST00000332772.4
|
SIMC1
|
SUMO-interacting motifs containing 1 |
| chr14_-_20801427 | 0.02 |
ENST00000557665.1
ENST00000358932.4 ENST00000353689.4 |
CCNB1IP1
|
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase |
| chr3_+_126113734 | 0.02 |
ENST00000352312.1
ENST00000393425.1 |
CCDC37
|
coiled-coil domain containing 37 |
| chr5_+_140743859 | 0.02 |
ENST00000518069.1
|
PCDHGA5
|
protocadherin gamma subfamily A, 5 |
| chrX_+_114874727 | 0.02 |
ENST00000543070.1
|
PLS3
|
plastin 3 |
| chr5_-_173043591 | 0.02 |
ENST00000285908.5
ENST00000480951.1 ENST00000311086.4 |
BOD1
|
biorientation of chromosomes in cell division 1 |
| chr10_-_75401500 | 0.02 |
ENST00000359322.4
|
MYOZ1
|
myozenin 1 |
| chr1_+_70876926 | 0.02 |
ENST00000370938.3
ENST00000346806.2 |
CTH
|
cystathionase (cystathionine gamma-lyase) |
| chr6_-_71666732 | 0.02 |
ENST00000230053.6
|
B3GAT2
|
beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) |
| chr2_+_102615416 | 0.02 |
ENST00000393414.2
|
IL1R2
|
interleukin 1 receptor, type II |
| chr9_-_104249400 | 0.01 |
ENST00000374848.3
|
TMEM246
|
transmembrane protein 246 |
| chr13_+_109248500 | 0.01 |
ENST00000356711.2
|
MYO16
|
myosin XVI |
| chr4_-_142134031 | 0.01 |
ENST00000420921.2
|
RNF150
|
ring finger protein 150 |
| chr1_+_46805832 | 0.01 |
ENST00000474844.1
|
NSUN4
|
NOP2/Sun domain family, member 4 |
| chr1_+_12806141 | 0.01 |
ENST00000288048.5
|
C1orf158
|
chromosome 1 open reading frame 158 |
| chr8_-_102181718 | 0.01 |
ENST00000565617.1
|
KB-1460A1.5
|
KB-1460A1.5 |
| chrX_+_129040122 | 0.01 |
ENST00000394422.3
ENST00000371051.5 |
UTP14A
|
UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) |
| chr12_-_110434183 | 0.01 |
ENST00000360185.4
ENST00000354574.4 ENST00000338373.5 ENST00000343646.5 ENST00000356259.4 ENST00000553118.1 |
GIT2
|
G protein-coupled receptor kinase interacting ArfGAP 2 |
| chr14_+_21249200 | 0.01 |
ENST00000304677.2
|
RNASE6
|
ribonuclease, RNase A family, k6 |
| chr19_+_3762645 | 0.01 |
ENST00000330133.4
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr7_-_105029812 | 0.01 |
ENST00000482897.1
|
SRPK2
|
SRSF protein kinase 2 |
| chr1_+_152975488 | 0.01 |
ENST00000542696.1
|
SPRR3
|
small proline-rich protein 3 |
| chr12_-_5849134 | 0.01 |
ENST00000545860.1
|
ANO2
|
anoctamin 2 |
| chr1_-_19578003 | 0.01 |
ENST00000375199.3
ENST00000375208.3 ENST00000356068.2 ENST00000477853.1 |
EMC1
|
ER membrane protein complex subunit 1 |
| chr11_-_104827425 | 0.01 |
ENST00000393150.3
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr4_+_26585538 | 0.01 |
ENST00000264866.4
|
TBC1D19
|
TBC1 domain family, member 19 |
| chrX_+_2976652 | 0.01 |
ENST00000537104.1
|
ARSF
|
arylsulfatase F |
| chr11_-_76381029 | 0.01 |
ENST00000407242.2
ENST00000421973.1 |
LRRC32
|
leucine rich repeat containing 32 |
| chr6_+_28249299 | 0.00 |
ENST00000405948.2
|
PGBD1
|
piggyBac transposable element derived 1 |
| chr11_-_76381781 | 0.00 |
ENST00000260061.5
ENST00000404995.1 |
LRRC32
|
leucine rich repeat containing 32 |
| chr9_-_2844058 | 0.00 |
ENST00000397885.2
|
KIAA0020
|
KIAA0020 |
| chrX_-_62974941 | 0.00 |
ENST00000374872.1
ENST00000253401.6 ENST00000374870.4 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr14_+_20811722 | 0.00 |
ENST00000429687.3
|
PARP2
|
poly (ADP-ribose) polymerase 2 |
| chrX_+_11129388 | 0.00 |
ENST00000321143.4
ENST00000380763.3 ENST00000380762.4 |
HCCS
|
holocytochrome c synthase |
| chr17_-_29624343 | 0.00 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.1 | GO:0031106 | septin ring assembly(GO:0000921) septin ring organization(GO:0031106) |
| 0.0 | 0.1 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.0 | 0.1 | GO:0018307 | enzyme active site formation(GO:0018307) |
| 0.0 | 0.4 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:0061083 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.1 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.0 | 0.1 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.1 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0032450 | alpha-1,4-glucosidase activity(GO:0004558) maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.1 | GO:0016295 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |