Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
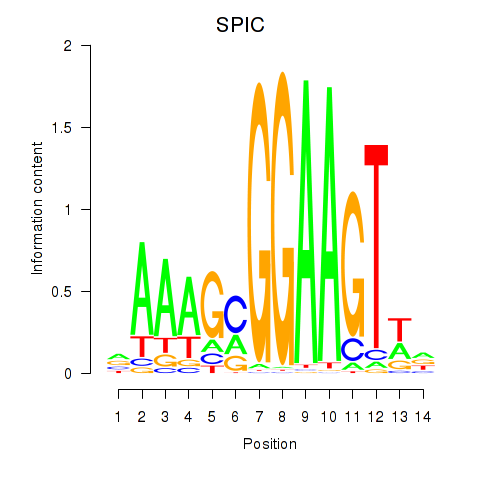
Results for SPIC
Z-value: 0.33
Transcription factors associated with SPIC
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SPIC
|
ENSG00000166211.6 | Spi-C transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SPIC | hg19_v2_chr12_+_101869096_101869199 | 0.41 | 4.2e-01 | Click! |
Activity profile of SPIC motif
Sorted Z-values of SPIC motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_108209897 | 0.21 |
ENST00000313516.5
|
THAP5
|
THAP domain containing 5 |
| chr2_-_40006289 | 0.19 |
ENST00000260619.6
ENST00000454352.2 |
THUMPD2
|
THUMP domain containing 2 |
| chr17_-_72709050 | 0.16 |
ENST00000583937.1
ENST00000301573.9 ENST00000326165.6 ENST00000469092.1 |
CD300LF
|
CD300 molecule-like family member f |
| chr19_+_45417504 | 0.16 |
ENST00000588750.1
ENST00000588802.1 |
APOC1
|
apolipoprotein C-I |
| chr9_-_5304432 | 0.15 |
ENST00000416837.1
ENST00000308420.3 |
RLN2
|
relaxin 2 |
| chr17_-_20370847 | 0.14 |
ENST00000423676.3
ENST00000324290.5 |
LGALS9B
|
lectin, galactoside-binding, soluble, 9B |
| chr4_-_74853897 | 0.14 |
ENST00000296028.3
|
PPBP
|
pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) |
| chr2_-_40006357 | 0.13 |
ENST00000505747.1
|
THUMPD2
|
THUMP domain containing 2 |
| chr5_-_82373260 | 0.12 |
ENST00000502346.1
|
TMEM167A
|
transmembrane protein 167A |
| chr7_-_91509972 | 0.12 |
ENST00000425936.1
|
MTERF
|
mitochondrial transcription termination factor |
| chr10_-_18948208 | 0.12 |
ENST00000607346.1
|
ARL5B-AS1
|
ARL5B antisense RNA 1 |
| chr1_+_110158726 | 0.12 |
ENST00000531734.1
|
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr1_+_219347186 | 0.12 |
ENST00000366928.5
|
LYPLAL1
|
lysophospholipase-like 1 |
| chr3_-_142720267 | 0.12 |
ENST00000597953.1
|
RP11-91G21.1
|
RP11-91G21.1 |
| chr11_-_18956556 | 0.12 |
ENST00000302797.3
|
MRGPRX1
|
MAS-related GPR, member X1 |
| chr12_+_21654714 | 0.11 |
ENST00000542038.1
ENST00000540141.1 ENST00000229314.5 |
GOLT1B
|
golgi transport 1B |
| chr5_+_115177178 | 0.11 |
ENST00000316788.7
|
AP3S1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chr15_-_33360342 | 0.11 |
ENST00000558197.1
|
FMN1
|
formin 1 |
| chr17_+_28443819 | 0.11 |
ENST00000479218.2
|
NSRP1
|
nuclear speckle splicing regulatory protein 1 |
| chr5_+_102455853 | 0.10 |
ENST00000515845.1
ENST00000321521.9 ENST00000507921.1 |
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr2_+_181988620 | 0.10 |
ENST00000428474.1
ENST00000424655.1 |
AC104820.2
|
AC104820.2 |
| chr3_-_28390298 | 0.10 |
ENST00000457172.1
|
AZI2
|
5-azacytidine induced 2 |
| chr20_+_34020827 | 0.10 |
ENST00000374375.1
|
GDF5OS
|
growth differentiation factor 5 opposite strand |
| chr3_+_149530836 | 0.10 |
ENST00000466478.1
ENST00000491086.1 ENST00000467977.1 |
RNF13
|
ring finger protein 13 |
| chr4_+_74702214 | 0.10 |
ENST00000226317.5
ENST00000515050.1 |
CXCL6
|
chemokine (C-X-C motif) ligand 6 |
| chr22_-_37880543 | 0.10 |
ENST00000442496.1
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chrX_+_24073048 | 0.10 |
ENST00000423068.1
|
EIF2S3
|
eukaryotic translation initiation factor 2, subunit 3 gamma, 52kDa |
| chr1_+_192127578 | 0.09 |
ENST00000367460.3
|
RGS18
|
regulator of G-protein signaling 18 |
| chr12_+_105724414 | 0.09 |
ENST00000443585.1
ENST00000552457.1 ENST00000549893.1 |
C12orf75
|
chromosome 12 open reading frame 75 |
| chr3_+_156544057 | 0.09 |
ENST00000498839.1
ENST00000470811.1 ENST00000356539.4 ENST00000483177.1 ENST00000477399.1 ENST00000491763.1 |
LEKR1
|
leucine, glutamate and lysine rich 1 |
| chr7_+_108210012 | 0.09 |
ENST00000249356.3
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9 |
| chr14_-_67826538 | 0.09 |
ENST00000553687.1
|
ATP6V1D
|
ATPase, H+ transporting, lysosomal 34kDa, V1 subunit D |
| chr1_+_112016414 | 0.09 |
ENST00000343534.5
ENST00000369718.3 |
C1orf162
|
chromosome 1 open reading frame 162 |
| chr3_-_28390415 | 0.08 |
ENST00000414162.1
ENST00000420543.2 |
AZI2
|
5-azacytidine induced 2 |
| chr19_-_1876156 | 0.08 |
ENST00000565797.1
|
CTB-31O20.2
|
CTB-31O20.2 |
| chr16_+_69373471 | 0.08 |
ENST00000569637.2
|
NIP7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr11_-_85780853 | 0.08 |
ENST00000531930.1
ENST00000528398.1 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr8_+_86019382 | 0.08 |
ENST00000360375.3
|
LRRCC1
|
leucine rich repeat and coiled-coil centrosomal protein 1 |
| chr3_-_49893958 | 0.08 |
ENST00000482243.1
ENST00000331456.2 ENST00000469027.1 |
TRAIP
|
TRAF interacting protein |
| chr1_+_120839005 | 0.08 |
ENST00000369390.3
ENST00000452190.1 |
FAM72B
|
family with sequence similarity 72, member B |
| chr6_+_139349817 | 0.08 |
ENST00000367660.3
|
ABRACL
|
ABRA C-terminal like |
| chr5_+_158690089 | 0.08 |
ENST00000296786.6
|
UBLCP1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr3_-_49893907 | 0.08 |
ENST00000482582.1
|
TRAIP
|
TRAF interacting protein |
| chr6_-_49430886 | 0.08 |
ENST00000274813.3
|
MUT
|
methylmalonyl CoA mutase |
| chr2_+_67624430 | 0.08 |
ENST00000272342.5
|
ETAA1
|
Ewing tumor-associated antigen 1 |
| chr15_+_63414760 | 0.08 |
ENST00000557972.1
|
LACTB
|
lactamase, beta |
| chr3_-_28390120 | 0.08 |
ENST00000334100.6
|
AZI2
|
5-azacytidine induced 2 |
| chr19_+_35939154 | 0.08 |
ENST00000599180.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr19_+_35940486 | 0.07 |
ENST00000246549.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr2_-_32264850 | 0.07 |
ENST00000295066.3
ENST00000342166.5 |
DPY30
|
dpy-30 homolog (C. elegans) |
| chr1_+_196788887 | 0.07 |
ENST00000320493.5
ENST00000367424.4 ENST00000367421.3 |
CFHR1
CFHR2
|
complement factor H-related 1 complement factor H-related 2 |
| chr5_+_162864575 | 0.07 |
ENST00000512163.1
ENST00000393929.1 ENST00000340828.2 ENST00000511683.2 ENST00000510097.1 ENST00000511490.2 ENST00000510664.1 |
CCNG1
|
cyclin G1 |
| chr15_+_63682335 | 0.07 |
ENST00000559379.1
ENST00000559821.1 |
RP11-321G12.1
|
RP11-321G12.1 |
| chrX_-_20159934 | 0.07 |
ENST00000379593.1
ENST00000379607.5 |
EIF1AX
|
eukaryotic translation initiation factor 1A, X-linked |
| chr4_-_112993808 | 0.07 |
ENST00000511219.1
|
RP11-269F21.3
|
RP11-269F21.3 |
| chr19_-_15344243 | 0.07 |
ENST00000602233.1
|
EPHX3
|
epoxide hydrolase 3 |
| chr12_-_110883346 | 0.07 |
ENST00000547365.1
|
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr13_+_43148281 | 0.07 |
ENST00000239849.6
ENST00000398795.2 ENST00000544862.1 |
TNFSF11
|
tumor necrosis factor (ligand) superfamily, member 11 |
| chr16_-_29465668 | 0.07 |
ENST00000569622.1
|
RP11-345J4.5
|
BolA-like protein 2 |
| chr1_-_71546690 | 0.07 |
ENST00000254821.6
|
ZRANB2
|
zinc finger, RAN-binding domain containing 2 |
| chr17_+_1959369 | 0.07 |
ENST00000576444.1
ENST00000322941.3 |
HIC1
|
hypermethylated in cancer 1 |
| chr12_-_44152551 | 0.07 |
ENST00000416848.2
ENST00000550784.1 ENST00000547156.1 ENST00000549868.1 ENST00000553166.1 ENST00000551923.1 ENST00000431332.3 ENST00000344862.5 |
PUS7L
|
pseudouridylate synthase 7 homolog (S. cerevisiae)-like |
| chr9_-_21995249 | 0.07 |
ENST00000494262.1
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A |
| chr5_+_59726565 | 0.07 |
ENST00000412930.2
|
FKSG52
|
FKSG52 |
| chr10_-_105156198 | 0.07 |
ENST00000369815.1
ENST00000309579.3 ENST00000337003.4 |
USMG5
|
up-regulated during skeletal muscle growth 5 homolog (mouse) |
| chr5_-_60458179 | 0.06 |
ENST00000507416.1
ENST00000339020.3 |
SMIM15
|
small integral membrane protein 15 |
| chr17_+_68047418 | 0.06 |
ENST00000586373.1
ENST00000588782.1 |
LINC01028
|
long intergenic non-protein coding RNA 1028 |
| chr13_+_41363581 | 0.06 |
ENST00000338625.4
|
SLC25A15
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15 |
| chr3_+_28390637 | 0.06 |
ENST00000420223.1
ENST00000383768.2 |
ZCWPW2
|
zinc finger, CW type with PWWP domain 2 |
| chr4_-_111120334 | 0.06 |
ENST00000503885.1
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr10_-_124713842 | 0.06 |
ENST00000481909.1
|
C10orf88
|
chromosome 10 open reading frame 88 |
| chr7_-_19157248 | 0.06 |
ENST00000242261.5
|
TWIST1
|
twist family bHLH transcription factor 1 |
| chr19_+_12780512 | 0.06 |
ENST00000242796.4
|
WDR83
|
WD repeat domain 83 |
| chr14_-_39639523 | 0.06 |
ENST00000330149.5
ENST00000554018.1 ENST00000347691.5 |
TRAPPC6B
|
trafficking protein particle complex 6B |
| chr8_-_101733794 | 0.06 |
ENST00000523555.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chrX_-_135056106 | 0.06 |
ENST00000433339.2
|
MMGT1
|
membrane magnesium transporter 1 |
| chr12_-_51785083 | 0.06 |
ENST00000603563.1
|
GALNT6
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) |
| chr2_+_138722028 | 0.06 |
ENST00000280096.5
|
HNMT
|
histamine N-methyltransferase |
| chr14_+_57735725 | 0.06 |
ENST00000431972.2
|
AP5M1
|
adaptor-related protein complex 5, mu 1 subunit |
| chr15_-_65282274 | 0.06 |
ENST00000204566.2
|
SPG21
|
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| chr3_-_52322019 | 0.06 |
ENST00000463624.1
|
WDR82
|
WD repeat domain 82 |
| chr10_-_98031265 | 0.06 |
ENST00000224337.5
ENST00000371176.2 |
BLNK
|
B-cell linker |
| chr17_-_60005365 | 0.06 |
ENST00000444766.3
|
INTS2
|
integrator complex subunit 2 |
| chr20_-_1569278 | 0.06 |
ENST00000262929.5
ENST00000567028.1 |
SIRPB1
RP4-576H24.4
|
signal-regulatory protein beta 1 Uncharacterized protein |
| chrX_-_135056216 | 0.06 |
ENST00000305963.2
|
MMGT1
|
membrane magnesium transporter 1 |
| chr3_+_101659682 | 0.06 |
ENST00000465215.1
|
RP11-221J22.1
|
RP11-221J22.1 |
| chr12_+_6833323 | 0.06 |
ENST00000544725.1
|
COPS7A
|
COP9 signalosome subunit 7A |
| chr2_+_113825547 | 0.06 |
ENST00000341010.2
ENST00000337569.3 |
IL1F10
|
interleukin 1 family, member 10 (theta) |
| chr19_+_21106081 | 0.06 |
ENST00000300540.3
ENST00000595854.1 ENST00000601284.1 ENST00000328178.8 ENST00000599885.1 ENST00000596476.1 ENST00000345030.6 |
ZNF85
|
zinc finger protein 85 |
| chr16_+_11439286 | 0.06 |
ENST00000312499.5
ENST00000576027.1 |
RMI2
|
RecQ mediated genome instability 2 |
| chr10_+_81891416 | 0.06 |
ENST00000372270.2
|
PLAC9
|
placenta-specific 9 |
| chr1_+_120839412 | 0.06 |
ENST00000355228.4
|
FAM72B
|
family with sequence similarity 72, member B |
| chr15_-_89089860 | 0.06 |
ENST00000558413.1
ENST00000564406.1 ENST00000268148.8 |
DET1
|
de-etiolated homolog 1 (Arabidopsis) |
| chr3_+_99979828 | 0.06 |
ENST00000485687.1
ENST00000344949.5 ENST00000394144.4 |
TBC1D23
|
TBC1 domain family, member 23 |
| chr3_-_47324008 | 0.06 |
ENST00000425853.1
|
KIF9
|
kinesin family member 9 |
| chr1_+_203830703 | 0.06 |
ENST00000414487.2
|
SNRPE
|
small nuclear ribonucleoprotein polypeptide E |
| chr12_-_15114492 | 0.06 |
ENST00000541546.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr16_+_23652773 | 0.06 |
ENST00000563998.1
ENST00000568589.1 ENST00000568272.1 |
DCTN5
|
dynactin 5 (p25) |
| chr16_+_69373323 | 0.06 |
ENST00000254940.5
|
NIP7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr20_+_2821366 | 0.05 |
ENST00000453689.1
ENST00000417508.1 |
VPS16
|
vacuolar protein sorting 16 homolog (S. cerevisiae) |
| chr4_-_84376953 | 0.05 |
ENST00000510985.1
ENST00000295488.3 |
HELQ
|
helicase, POLQ-like |
| chr14_+_57735614 | 0.05 |
ENST00000261558.3
|
AP5M1
|
adaptor-related protein complex 5, mu 1 subunit |
| chr3_-_156272924 | 0.05 |
ENST00000467789.1
ENST00000265044.2 |
SSR3
|
signal sequence receptor, gamma (translocon-associated protein gamma) |
| chr8_+_42911552 | 0.05 |
ENST00000525699.1
ENST00000529687.1 |
FNTA
|
farnesyltransferase, CAAX box, alpha |
| chr1_+_206138457 | 0.05 |
ENST00000367128.3
ENST00000431655.2 |
FAM72A
|
family with sequence similarity 72, member A |
| chr12_-_120663792 | 0.05 |
ENST00000546532.1
ENST00000548912.1 |
PXN
|
paxillin |
| chr17_-_29641104 | 0.05 |
ENST00000577894.1
ENST00000330927.4 |
EVI2B
|
ecotropic viral integration site 2B |
| chrX_+_51075658 | 0.05 |
ENST00000356450.2
|
NUDT10
|
nudix (nucleoside diphosphate linked moiety X)-type motif 10 |
| chr16_-_31085514 | 0.05 |
ENST00000300849.4
|
ZNF668
|
zinc finger protein 668 |
| chr15_-_80263506 | 0.05 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr17_+_79633745 | 0.05 |
ENST00000574107.1
|
CCDC137
|
coiled-coil domain containing 137 |
| chrX_+_24072833 | 0.05 |
ENST00000253039.4
|
EIF2S3
|
eukaryotic translation initiation factor 2, subunit 3 gamma, 52kDa |
| chr6_+_26183958 | 0.05 |
ENST00000356530.3
|
HIST1H2BE
|
histone cluster 1, H2be |
| chr17_+_18684563 | 0.05 |
ENST00000476139.1
|
TVP23B
|
trans-golgi network vesicle protein 23 homolog B (S. cerevisiae) |
| chr19_+_55084438 | 0.05 |
ENST00000439534.1
|
LILRA2
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 2 |
| chr8_+_104426942 | 0.05 |
ENST00000297579.5
|
DCAF13
|
DDB1 and CUL4 associated factor 13 |
| chr20_-_1569351 | 0.05 |
ENST00000563840.1
|
SIRPB1
|
signal-regulatory protein beta 1 |
| chr11_-_22647350 | 0.05 |
ENST00000327470.3
|
FANCF
|
Fanconi anemia, complementation group F |
| chr4_+_41992489 | 0.05 |
ENST00000264451.7
|
SLC30A9
|
solute carrier family 30 (zinc transporter), member 9 |
| chr6_-_74019870 | 0.05 |
ENST00000370384.3
|
KHDC1
|
KH homology domain containing 1 |
| chr14_-_57735528 | 0.05 |
ENST00000340918.7
ENST00000413566.2 |
EXOC5
|
exocyst complex component 5 |
| chr5_-_114632307 | 0.05 |
ENST00000506442.1
ENST00000379611.5 |
CCDC112
|
coiled-coil domain containing 112 |
| chr5_+_110074685 | 0.05 |
ENST00000355943.3
ENST00000447245.2 |
SLC25A46
|
solute carrier family 25, member 46 |
| chr8_-_104427289 | 0.05 |
ENST00000543107.1
|
SLC25A32
|
solute carrier family 25 (mitochondrial folate carrier), member 32 |
| chr19_-_10446449 | 0.05 |
ENST00000592439.1
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr9_+_74526532 | 0.05 |
ENST00000486911.2
|
C9orf85
|
chromosome 9 open reading frame 85 |
| chr5_+_134181625 | 0.05 |
ENST00000394976.3
|
C5orf24
|
chromosome 5 open reading frame 24 |
| chr14_-_58893876 | 0.05 |
ENST00000555097.1
ENST00000555404.1 |
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr10_-_96829246 | 0.05 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr6_-_11779014 | 0.05 |
ENST00000229583.5
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr11_+_124824000 | 0.05 |
ENST00000529051.1
ENST00000344762.5 |
CCDC15
|
coiled-coil domain containing 15 |
| chr17_+_18380051 | 0.04 |
ENST00000581545.1
ENST00000582333.1 ENST00000328114.6 ENST00000412421.2 ENST00000583322.1 ENST00000584941.1 |
LGALS9C
|
lectin, galactoside-binding, soluble, 9C |
| chr14_+_63671105 | 0.04 |
ENST00000316754.3
|
RHOJ
|
ras homolog family member J |
| chr8_+_38662960 | 0.04 |
ENST00000524193.1
|
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr4_-_88312301 | 0.04 |
ENST00000507286.1
|
HSD17B11
|
hydroxysteroid (17-beta) dehydrogenase 11 |
| chr12_+_6833237 | 0.04 |
ENST00000229251.3
ENST00000539735.1 ENST00000538410.1 |
COPS7A
|
COP9 signalosome subunit 7A |
| chr19_-_41903161 | 0.04 |
ENST00000602129.1
ENST00000593771.1 ENST00000596905.1 ENST00000221233.4 |
EXOSC5
|
exosome component 5 |
| chr2_+_201981527 | 0.04 |
ENST00000441224.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr6_-_117747015 | 0.04 |
ENST00000368508.3
ENST00000368507.3 |
ROS1
|
c-ros oncogene 1 , receptor tyrosine kinase |
| chr7_+_23145347 | 0.04 |
ENST00000322231.7
|
KLHL7
|
kelch-like family member 7 |
| chr3_-_179322436 | 0.04 |
ENST00000392659.2
ENST00000476781.1 |
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr1_-_246729544 | 0.04 |
ENST00000544618.1
ENST00000366514.4 |
TFB2M
|
transcription factor B2, mitochondrial |
| chr7_-_108210048 | 0.04 |
ENST00000415914.3
ENST00000438865.1 |
THAP5
|
THAP domain containing 5 |
| chrX_+_123095860 | 0.04 |
ENST00000428941.1
|
STAG2
|
stromal antigen 2 |
| chr12_+_44152740 | 0.04 |
ENST00000440781.2
ENST00000431837.1 ENST00000550616.1 ENST00000448290.2 ENST00000551736.1 |
IRAK4
|
interleukin-1 receptor-associated kinase 4 |
| chr19_-_40596767 | 0.04 |
ENST00000599972.1
ENST00000450241.2 ENST00000595687.2 |
ZNF780A
|
zinc finger protein 780A |
| chr12_-_49523896 | 0.04 |
ENST00000549870.1
|
TUBA1B
|
tubulin, alpha 1b |
| chr5_-_142782862 | 0.04 |
ENST00000415690.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr18_+_55862873 | 0.04 |
ENST00000588494.1
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr5_-_148929848 | 0.04 |
ENST00000504676.1
ENST00000515435.1 |
CSNK1A1
|
casein kinase 1, alpha 1 |
| chr8_+_18067602 | 0.04 |
ENST00000307719.4
ENST00000545197.1 ENST00000539092.1 ENST00000541942.1 ENST00000518029.1 |
NAT1
|
N-acetyltransferase 1 (arylamine N-acetyltransferase) |
| chr6_-_33239712 | 0.04 |
ENST00000436044.2
|
VPS52
|
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr2_-_37458749 | 0.04 |
ENST00000234170.5
|
CEBPZ
|
CCAAT/enhancer binding protein (C/EBP), zeta |
| chr10_+_91461337 | 0.04 |
ENST00000260753.4
ENST00000416354.1 ENST00000394289.2 ENST00000371728.3 |
KIF20B
|
kinesin family member 20B |
| chr12_+_53693466 | 0.04 |
ENST00000267103.5
ENST00000548632.1 |
C12orf10
|
chromosome 12 open reading frame 10 |
| chr21_+_17214724 | 0.04 |
ENST00000449491.1
|
USP25
|
ubiquitin specific peptidase 25 |
| chr7_-_32529973 | 0.04 |
ENST00000410044.1
ENST00000409987.1 ENST00000409782.1 ENST00000450169.2 |
LSM5
|
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr1_-_157789850 | 0.04 |
ENST00000491942.1
ENST00000358292.3 ENST00000368176.3 |
FCRL1
|
Fc receptor-like 1 |
| chr16_-_23652570 | 0.04 |
ENST00000261584.4
|
PALB2
|
partner and localizer of BRCA2 |
| chr11_+_112047087 | 0.04 |
ENST00000526088.1
ENST00000532593.1 ENST00000531169.1 |
BCO2
|
beta-carotene oxygenase 2 |
| chr11_+_327171 | 0.04 |
ENST00000534483.1
ENST00000524824.1 ENST00000531076.1 |
RP11-326C3.12
|
RP11-326C3.12 |
| chr5_-_149465990 | 0.04 |
ENST00000543093.1
|
CSF1R
|
colony stimulating factor 1 receptor |
| chr1_+_222886694 | 0.04 |
ENST00000426638.1
ENST00000537020.1 ENST00000539697.1 |
BROX
|
BRO1 domain and CAAX motif containing |
| chr15_+_90735145 | 0.04 |
ENST00000559792.1
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr1_-_222886526 | 0.04 |
ENST00000541237.1
|
AIDA
|
axin interactor, dorsalization associated |
| chr4_-_99578776 | 0.04 |
ENST00000515287.1
|
TSPAN5
|
tetraspanin 5 |
| chr11_-_61101247 | 0.04 |
ENST00000543627.1
|
DDB1
|
damage-specific DNA binding protein 1, 127kDa |
| chr1_+_206138884 | 0.04 |
ENST00000341209.5
ENST00000607379.1 |
FAM72A
|
family with sequence similarity 72, member A |
| chrX_+_55478538 | 0.04 |
ENST00000342972.1
|
MAGEH1
|
melanoma antigen family H, 1 |
| chr7_-_5553369 | 0.04 |
ENST00000453700.3
ENST00000382368.3 |
FBXL18
|
F-box and leucine-rich repeat protein 18 |
| chr6_+_151773583 | 0.04 |
ENST00000545879.1
|
C6orf211
|
chromosome 6 open reading frame 211 |
| chrX_+_100645812 | 0.04 |
ENST00000427805.2
ENST00000553110.3 ENST00000392994.3 ENST00000409338.1 ENST00000409170.3 |
RPL36A
RPL36A-HNRNPH2
|
ribosomal protein L36a RPL36A-HNRNPH2 readthrough |
| chr19_+_6740888 | 0.04 |
ENST00000596673.1
|
TRIP10
|
thyroid hormone receptor interactor 10 |
| chr6_-_100016527 | 0.04 |
ENST00000523985.1
ENST00000518714.1 ENST00000520371.1 |
CCNC
|
cyclin C |
| chr17_+_21730180 | 0.04 |
ENST00000584398.1
|
UBBP4
|
ubiquitin B pseudogene 4 |
| chr7_+_95401877 | 0.04 |
ENST00000524053.1
ENST00000324972.6 ENST00000537881.1 ENST00000437599.1 ENST00000359388.4 ENST00000413338.1 |
DYNC1I1
|
dynein, cytoplasmic 1, intermediate chain 1 |
| chr6_-_32557610 | 0.04 |
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1 |
| chr3_-_196014520 | 0.04 |
ENST00000441879.1
ENST00000292823.2 ENST00000411591.1 ENST00000431016.1 ENST00000443555.1 |
PCYT1A
|
phosphate cytidylyltransferase 1, choline, alpha |
| chr1_-_149859466 | 0.04 |
ENST00000331128.3
|
HIST2H2AB
|
histone cluster 2, H2ab |
| chr12_+_120884222 | 0.04 |
ENST00000551765.1
ENST00000229384.5 |
GATC
|
glutamyl-tRNA(Gln) amidotransferase, subunit C |
| chr19_-_50464338 | 0.04 |
ENST00000426971.2
ENST00000447370.2 |
SIGLEC11
|
sialic acid binding Ig-like lectin 11 |
| chr22_-_30942669 | 0.04 |
ENST00000402034.2
|
SEC14L6
|
SEC14-like 6 (S. cerevisiae) |
| chr1_-_108742957 | 0.04 |
ENST00000565488.1
|
SLC25A24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr6_-_37467628 | 0.04 |
ENST00000373408.3
|
CCDC167
|
coiled-coil domain containing 167 |
| chr8_+_104427581 | 0.04 |
ENST00000521716.1
ENST00000521971.1 ENST00000519682.1 |
DCAF13
|
DDB1 and CUL4 associated factor 13 |
| chr17_-_15168624 | 0.04 |
ENST00000312280.3
ENST00000494511.1 ENST00000580584.1 |
PMP22
|
peripheral myelin protein 22 |
| chr3_-_46786245 | 0.04 |
ENST00000442359.2
|
PRSS45
|
protease, serine, 45 |
| chr1_-_53833812 | 0.04 |
ENST00000449958.1
|
RP11-117D22.2
|
RP11-117D22.2 |
| chr14_+_58894404 | 0.04 |
ENST00000554463.1
ENST00000555833.1 |
KIAA0586
|
KIAA0586 |
| chr6_+_142623758 | 0.04 |
ENST00000541199.1
ENST00000435011.2 |
GPR126
|
G protein-coupled receptor 126 |
| chr19_-_37663572 | 0.04 |
ENST00000588354.1
ENST00000292841.5 ENST00000355533.2 ENST00000356958.4 |
ZNF585A
|
zinc finger protein 585A |
| chr4_+_87857538 | 0.04 |
ENST00000511442.1
|
AFF1
|
AF4/FMR2 family, member 1 |
| chr1_-_211848899 | 0.04 |
ENST00000366998.3
ENST00000540251.1 ENST00000366999.4 |
NEK2
|
NIMA-related kinase 2 |
| chr2_-_3381561 | 0.04 |
ENST00000443925.2
ENST00000441271.1 ENST00000444776.1 ENST00000398659.4 ENST00000382125.4 |
TSSC1
|
tumor suppressing subtransferable candidate 1 |
| chr2_+_181988560 | 0.04 |
ENST00000424170.1
ENST00000435411.1 |
AC104820.2
|
AC104820.2 |
| chr8_-_101962777 | 0.04 |
ENST00000395951.3
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr11_-_96076334 | 0.04 |
ENST00000524717.1
|
MAML2
|
mastermind-like 2 (Drosophila) |
| chr16_+_23652700 | 0.04 |
ENST00000300087.2
|
DCTN5
|
dynactin 5 (p25) |
| chr5_-_132362226 | 0.04 |
ENST00000509437.1
ENST00000355372.2 ENST00000513541.1 ENST00000509008.1 ENST00000513848.1 ENST00000504170.1 ENST00000324170.3 |
ZCCHC10
|
zinc finger, CCHC domain containing 10 |
| chr18_+_6256746 | 0.04 |
ENST00000578427.1
|
RP11-760N9.1
|
RP11-760N9.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SPIC
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1902214 | regulation of interleukin-4-mediated signaling pathway(GO:1902214) |
| 0.1 | 0.2 | GO:0002525 | acute inflammatory response to non-antigenic stimulus(GO:0002525) regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 0.0 | 0.2 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.0 | 0.3 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.1 | GO:0071848 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 0.0 | 0.1 | GO:2000276 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.0 | 0.1 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.0 | GO:1902954 | regulation of early endosome to recycling endosome transport(GO:1902954) |
| 0.0 | 0.0 | GO:0070902 | mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.0 | GO:0042214 | terpene metabolic process(GO:0042214) |
| 0.0 | 0.0 | GO:0002537 | nitric oxide production involved in inflammatory response(GO:0002537) |
| 0.0 | 0.0 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.0 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.0 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.0 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.0 | GO:0036257 | cell separation after cytokinesis(GO:0000920) multivesicular body organization(GO:0036257) multivesicular body assembly(GO:0036258) viral budding via host ESCRT complex(GO:0039702) viral budding(GO:0046755) multi-organism organelle organization(GO:1902590) multi-organism membrane budding(GO:1902592) ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.0 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.0 | 0.0 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.0 | 0.0 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.0 | 0.0 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.1 | GO:0006404 | RNA import into nucleus(GO:0006404) |
| 0.0 | 0.1 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 0.0 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.0 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.0 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.0 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.0 | 0.0 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.1 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.0 | 0.2 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.0 | GO:0004040 | amidase activity(GO:0004040) |
| 0.0 | 0.0 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.3 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.0 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.0 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.0 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.0 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.0 | GO:0034038 | deoxyhypusine synthase activity(GO:0034038) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |