Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
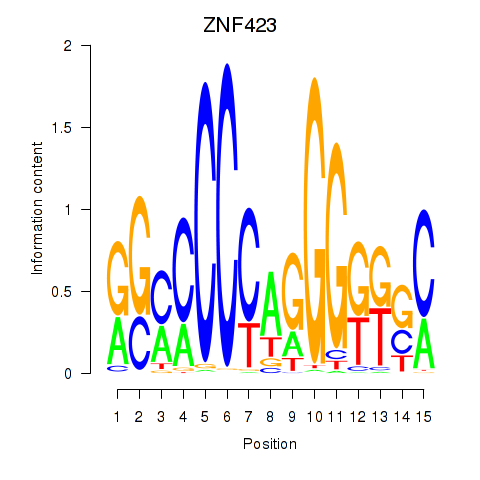
Results for ZNF423
Z-value: 0.32
Transcription factors associated with ZNF423
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF423
|
ENSG00000102935.7 | zinc finger protein 423 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF423 | hg19_v2_chr16_-_49891694_49891830 | -0.72 | 1.1e-01 | Click! |
Activity profile of ZNF423 motif
Sorted Z-values of ZNF423 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_77020325 | 0.22 |
ENST00000311661.4
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr17_+_77020224 | 0.22 |
ENST00000339142.2
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr15_-_41522889 | 0.18 |
ENST00000458580.2
ENST00000314992.5 ENST00000558396.1 |
EXD1
|
exonuclease 3'-5' domain containing 1 |
| chr17_+_6347761 | 0.17 |
ENST00000250056.8
ENST00000571373.1 ENST00000570337.2 ENST00000572595.2 ENST00000576056.1 |
FAM64A
|
family with sequence similarity 64, member A |
| chr11_-_44972299 | 0.16 |
ENST00000528473.1
|
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr4_+_74735102 | 0.15 |
ENST00000395761.3
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
| chr7_+_150748288 | 0.15 |
ENST00000490540.1
|
ASIC3
|
acid-sensing (proton-gated) ion channel 3 |
| chr20_-_36794902 | 0.14 |
ENST00000373403.3
|
TGM2
|
transglutaminase 2 |
| chr17_+_77020146 | 0.14 |
ENST00000579760.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr11_-_44972418 | 0.12 |
ENST00000525680.1
ENST00000528290.1 ENST00000530035.1 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr20_-_36794938 | 0.12 |
ENST00000453095.1
|
TGM2
|
transglutaminase 2 |
| chr11_+_60691924 | 0.11 |
ENST00000544065.1
ENST00000453848.2 ENST00000005286.4 |
TMEM132A
|
transmembrane protein 132A |
| chr12_-_50101165 | 0.11 |
ENST00000352151.5
ENST00000335154.5 ENST00000293590.5 |
FMNL3
|
formin-like 3 |
| chr17_-_79212825 | 0.10 |
ENST00000374769.2
|
ENTHD2
|
ENTH domain containing 2 |
| chr6_-_31080336 | 0.10 |
ENST00000259870.3
|
C6orf15
|
chromosome 6 open reading frame 15 |
| chr11_-_61658853 | 0.09 |
ENST00000525588.1
ENST00000540820.1 |
FADS3
|
fatty acid desaturase 3 |
| chr12_-_50101003 | 0.09 |
ENST00000550488.1
|
FMNL3
|
formin-like 3 |
| chr11_-_61659006 | 0.08 |
ENST00000278829.2
|
FADS3
|
fatty acid desaturase 3 |
| chr17_+_6347729 | 0.08 |
ENST00000572447.1
|
FAM64A
|
family with sequence similarity 64, member A |
| chr7_-_150754935 | 0.08 |
ENST00000297518.4
|
CDK5
|
cyclin-dependent kinase 5 |
| chr9_+_116638630 | 0.08 |
ENST00000452710.1
ENST00000374124.4 |
ZNF618
|
zinc finger protein 618 |
| chr9_-_131486367 | 0.08 |
ENST00000372663.4
ENST00000406904.2 ENST00000452105.1 ENST00000372672.2 ENST00000372667.5 |
ZDHHC12
|
zinc finger, DHHC-type containing 12 |
| chr22_+_35653445 | 0.07 |
ENST00000420166.1
ENST00000444518.2 ENST00000455359.1 ENST00000216106.5 |
HMGXB4
|
HMG box domain containing 4 |
| chr11_-_44971702 | 0.07 |
ENST00000533940.1
ENST00000533937.1 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr11_-_64013663 | 0.07 |
ENST00000392210.2
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr1_+_155278625 | 0.07 |
ENST00000368356.4
ENST00000356657.6 |
FDPS
|
farnesyl diphosphate synthase |
| chr19_+_45971246 | 0.07 |
ENST00000585836.1
ENST00000417353.2 ENST00000353609.3 ENST00000591858.1 ENST00000443841.2 ENST00000590335.1 |
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr6_-_32160622 | 0.07 |
ENST00000487761.1
ENST00000375040.3 |
GPSM3
|
G-protein signaling modulator 3 |
| chr15_-_75660919 | 0.07 |
ENST00000569482.1
ENST00000565683.1 ENST00000561615.1 ENST00000563622.1 ENST00000568374.1 ENST00000566256.1 ENST00000267978.5 |
MAN2C1
|
mannosidase, alpha, class 2C, member 1 |
| chr20_+_44462430 | 0.07 |
ENST00000462307.1
|
SNX21
|
sorting nexin family member 21 |
| chr17_-_1029052 | 0.07 |
ENST00000574437.1
|
ABR
|
active BCR-related |
| chr9_+_116638562 | 0.07 |
ENST00000374126.5
ENST00000288466.7 |
ZNF618
|
zinc finger protein 618 |
| chr6_-_91296737 | 0.07 |
ENST00000369332.3
ENST00000369329.3 |
MAP3K7
|
mitogen-activated protein kinase kinase kinase 7 |
| chr4_+_152330409 | 0.06 |
ENST00000513086.1
|
FAM160A1
|
family with sequence similarity 160, member A1 |
| chr19_+_20011744 | 0.06 |
ENST00000588146.1
|
ZNF93
|
zinc finger protein 93 |
| chr17_-_27230035 | 0.06 |
ENST00000378895.4
ENST00000394901.3 |
DHRS13
|
dehydrogenase/reductase (SDR family) member 13 |
| chr22_+_39853258 | 0.06 |
ENST00000341184.6
|
MGAT3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase |
| chr9_-_16870704 | 0.06 |
ENST00000380672.4
ENST00000380667.2 ENST00000380666.2 ENST00000486514.1 |
BNC2
|
basonuclin 2 |
| chr11_-_44972476 | 0.06 |
ENST00000527685.1
ENST00000308212.5 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr20_+_44462749 | 0.06 |
ENST00000372541.1
|
SNX21
|
sorting nexin family member 21 |
| chr16_+_30675654 | 0.06 |
ENST00000287468.5
ENST00000395073.2 |
FBRS
|
fibrosin |
| chr8_+_22022800 | 0.06 |
ENST00000397814.3
|
BMP1
|
bone morphogenetic protein 1 |
| chr1_+_17914907 | 0.06 |
ENST00000375420.3
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr16_+_27324983 | 0.05 |
ENST00000566117.1
|
IL4R
|
interleukin 4 receptor |
| chr22_-_38484922 | 0.05 |
ENST00000428572.1
|
BAIAP2L2
|
BAI1-associated protein 2-like 2 |
| chr6_-_91296602 | 0.05 |
ENST00000369325.3
ENST00000369327.3 |
MAP3K7
|
mitogen-activated protein kinase kinase kinase 7 |
| chr7_-_5570229 | 0.05 |
ENST00000331789.5
|
ACTB
|
actin, beta |
| chr5_+_141303461 | 0.05 |
ENST00000508751.1
|
KIAA0141
|
KIAA0141 |
| chr6_+_33589161 | 0.05 |
ENST00000605930.1
|
ITPR3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr10_-_121296045 | 0.04 |
ENST00000392865.1
|
RGS10
|
regulator of G-protein signaling 10 |
| chr21_-_18985158 | 0.04 |
ENST00000339775.6
|
BTG3
|
BTG family, member 3 |
| chr11_-_504489 | 0.04 |
ENST00000529368.1
|
RNH1
|
ribonuclease/angiogenin inhibitor 1 |
| chr5_+_154181816 | 0.04 |
ENST00000518677.1
|
LARP1
|
La ribonucleoprotein domain family, member 1 |
| chr19_-_10450287 | 0.04 |
ENST00000589261.1
ENST00000590569.1 ENST00000589580.1 ENST00000589249.1 |
ICAM3
|
intercellular adhesion molecule 3 |
| chr2_+_219125714 | 0.04 |
ENST00000522678.1
ENST00000519574.1 ENST00000521462.1 |
GPBAR1
|
G protein-coupled bile acid receptor 1 |
| chr2_-_206950996 | 0.04 |
ENST00000414320.1
|
INO80D
|
INO80 complex subunit D |
| chr19_-_10679644 | 0.04 |
ENST00000393599.2
|
CDKN2D
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr2_+_171785012 | 0.04 |
ENST00000234160.4
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa |
| chr19_-_36391434 | 0.04 |
ENST00000396901.1
ENST00000585925.1 |
NFKBID
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, delta |
| chr3_-_48594248 | 0.04 |
ENST00000545984.1
ENST00000232375.3 ENST00000416568.1 ENST00000383734.2 ENST00000541519.1 ENST00000412035.1 |
PFKFB4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr12_+_7037461 | 0.04 |
ENST00000396684.2
|
ATN1
|
atrophin 1 |
| chr13_+_96743093 | 0.04 |
ENST00000376705.2
|
HS6ST3
|
heparan sulfate 6-O-sulfotransferase 3 |
| chrX_-_51239425 | 0.04 |
ENST00000375992.3
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chr5_-_176057365 | 0.04 |
ENST00000310112.3
|
SNCB
|
synuclein, beta |
| chr11_+_73019282 | 0.04 |
ENST00000263674.3
|
ARHGEF17
|
Rho guanine nucleotide exchange factor (GEF) 17 |
| chr12_+_7342271 | 0.04 |
ENST00000434354.2
ENST00000544456.1 ENST00000545574.1 ENST00000420616.2 |
PEX5
|
peroxisomal biogenesis factor 5 |
| chr1_+_1222489 | 0.04 |
ENST00000379099.3
|
SCNN1D
|
sodium channel, non-voltage-gated 1, delta subunit |
| chr19_+_41770349 | 0.03 |
ENST00000602130.1
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr15_+_41523417 | 0.03 |
ENST00000560397.1
|
CHP1
|
calcineurin-like EF-hand protein 1 |
| chr9_-_130659679 | 0.03 |
ENST00000373141.1
|
ST6GALNAC6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr21_+_45432174 | 0.03 |
ENST00000380221.3
ENST00000291574.4 |
TRAPPC10
|
trafficking protein particle complex 10 |
| chr1_+_10459111 | 0.03 |
ENST00000541529.1
ENST00000270776.8 ENST00000483936.1 ENST00000538557.1 |
PGD
|
phosphogluconate dehydrogenase |
| chr6_-_143266297 | 0.03 |
ENST00000367603.2
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2 |
| chr7_+_20370300 | 0.03 |
ENST00000537992.1
|
ITGB8
|
integrin, beta 8 |
| chrX_+_136648297 | 0.03 |
ENST00000287538.5
|
ZIC3
|
Zic family member 3 |
| chr19_-_10450328 | 0.03 |
ENST00000160262.5
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr10_+_124914285 | 0.03 |
ENST00000407911.2
|
BUB3
|
BUB3 mitotic checkpoint protein |
| chr8_+_22022223 | 0.03 |
ENST00000306385.5
|
BMP1
|
bone morphogenetic protein 1 |
| chr3_+_9958870 | 0.03 |
ENST00000413608.1
|
IL17RC
|
interleukin 17 receptor C |
| chr14_-_93582214 | 0.03 |
ENST00000556603.2
ENST00000354313.3 |
ITPK1
|
inositol-tetrakisphosphate 1-kinase |
| chr6_+_42896865 | 0.03 |
ENST00000372836.4
ENST00000394142.3 |
CNPY3
|
canopy FGF signaling regulator 3 |
| chr7_-_27219632 | 0.03 |
ENST00000470747.4
|
RP1-170O19.20
|
Uncharacterized protein |
| chr7_+_20370746 | 0.03 |
ENST00000222573.4
|
ITGB8
|
integrin, beta 8 |
| chr6_+_151186554 | 0.03 |
ENST00000367321.3
ENST00000367307.4 |
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr8_+_22022653 | 0.03 |
ENST00000354870.5
ENST00000397816.3 ENST00000306349.8 |
BMP1
|
bone morphogenetic protein 1 |
| chr11_-_67120974 | 0.03 |
ENST00000539074.1
ENST00000312419.3 |
POLD4
|
polymerase (DNA-directed), delta 4, accessory subunit |
| chr22_-_43045574 | 0.03 |
ENST00000352397.5
|
CYB5R3
|
cytochrome b5 reductase 3 |
| chr2_+_171785824 | 0.03 |
ENST00000452526.2
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa |
| chr9_-_130659569 | 0.03 |
ENST00000542456.1
|
ST6GALNAC6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr10_+_124913930 | 0.02 |
ENST00000368858.5
|
BUB3
|
BUB3 mitotic checkpoint protein |
| chr14_-_93582148 | 0.02 |
ENST00000267615.6
ENST00000553452.1 |
ITPK1
|
inositol-tetrakisphosphate 1-kinase |
| chr13_-_46626820 | 0.02 |
ENST00000428921.1
|
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr17_-_48227877 | 0.02 |
ENST00000316878.6
|
PPP1R9B
|
protein phosphatase 1, regulatory subunit 9B |
| chr4_-_20985632 | 0.02 |
ENST00000359001.5
|
KCNIP4
|
Kv channel interacting protein 4 |
| chr11_+_61595752 | 0.02 |
ENST00000521849.1
|
FADS2
|
fatty acid desaturase 2 |
| chr1_-_48462566 | 0.02 |
ENST00000606738.2
|
TRABD2B
|
TraB domain containing 2B |
| chr19_+_41770269 | 0.02 |
ENST00000378215.4
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr3_-_195808980 | 0.02 |
ENST00000360110.4
|
TFRC
|
transferrin receptor |
| chr12_-_125348448 | 0.02 |
ENST00000339570.5
|
SCARB1
|
scavenger receptor class B, member 1 |
| chrX_+_136648643 | 0.02 |
ENST00000370606.3
|
ZIC3
|
Zic family member 3 |
| chr8_-_42234745 | 0.02 |
ENST00000220812.2
|
DKK4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chr12_+_7342441 | 0.02 |
ENST00000412720.2
ENST00000396637.3 |
PEX5
|
peroxisomal biogenesis factor 5 |
| chr11_+_61595572 | 0.02 |
ENST00000517312.1
|
FADS2
|
fatty acid desaturase 2 |
| chr6_+_168841817 | 0.02 |
ENST00000356284.2
ENST00000354536.5 |
SMOC2
|
SPARC related modular calcium binding 2 |
| chr1_-_155177677 | 0.02 |
ENST00000368378.3
ENST00000541990.1 ENST00000457183.2 |
THBS3
|
thrombospondin 3 |
| chr21_-_18985230 | 0.02 |
ENST00000457956.1
ENST00000348354.6 |
BTG3
|
BTG family, member 3 |
| chr8_-_133493200 | 0.02 |
ENST00000388996.4
|
KCNQ3
|
potassium voltage-gated channel, KQT-like subfamily, member 3 |
| chr20_-_45976816 | 0.02 |
ENST00000441977.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr3_+_9958758 | 0.02 |
ENST00000383812.4
ENST00000438091.1 ENST00000295981.3 ENST00000436503.1 ENST00000403601.3 ENST00000416074.2 ENST00000455057.1 |
IL17RC
|
interleukin 17 receptor C |
| chr19_+_6740888 | 0.02 |
ENST00000596673.1
|
TRIP10
|
thyroid hormone receptor interactor 10 |
| chr19_-_57183114 | 0.02 |
ENST00000537055.2
ENST00000601659.1 |
ZNF835
|
zinc finger protein 835 |
| chr19_-_15311713 | 0.02 |
ENST00000601011.1
ENST00000263388.2 |
NOTCH3
|
notch 3 |
| chr12_+_54410664 | 0.02 |
ENST00000303406.4
|
HOXC4
|
homeobox C4 |
| chr1_+_55271736 | 0.02 |
ENST00000358193.3
ENST00000371273.3 |
C1orf177
|
chromosome 1 open reading frame 177 |
| chr1_-_204121013 | 0.02 |
ENST00000367201.3
|
ETNK2
|
ethanolamine kinase 2 |
| chr3_+_180630090 | 0.02 |
ENST00000357559.4
ENST00000305586.7 |
FXR1
|
fragile X mental retardation, autosomal homolog 1 |
| chr16_+_27325202 | 0.01 |
ENST00000395762.2
ENST00000562142.1 ENST00000561742.1 ENST00000543915.2 ENST00000449195.1 ENST00000380922.3 ENST00000563002.1 |
IL4R
|
interleukin 4 receptor |
| chr16_+_71392616 | 0.01 |
ENST00000349553.5
ENST00000302628.4 ENST00000562305.1 |
CALB2
|
calbindin 2 |
| chr9_+_112542572 | 0.01 |
ENST00000374530.3
|
PALM2-AKAP2
|
PALM2-AKAP2 readthrough |
| chr11_-_47207950 | 0.01 |
ENST00000298838.6
ENST00000531226.1 ENST00000524509.1 ENST00000528201.1 ENST00000530513.1 |
PACSIN3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr1_-_165414414 | 0.01 |
ENST00000359842.5
|
RXRG
|
retinoid X receptor, gamma |
| chr6_-_35480640 | 0.01 |
ENST00000428978.1
ENST00000322263.4 |
TULP1
|
tubby like protein 1 |
| chr19_+_45582453 | 0.01 |
ENST00000591607.1
ENST00000591747.1 ENST00000270257.4 ENST00000391951.2 ENST00000587566.1 |
GEMIN7
MARK4
|
gem (nuclear organelle) associated protein 7 MAP/microtubule affinity-regulating kinase 4 |
| chr4_+_128982490 | 0.01 |
ENST00000394288.3
ENST00000432347.2 ENST00000264584.5 ENST00000441387.1 ENST00000427266.1 ENST00000354456.3 |
LARP1B
|
La ribonucleoprotein domain family, member 1B |
| chr1_-_33366931 | 0.01 |
ENST00000373463.3
ENST00000329151.5 |
TMEM54
|
transmembrane protein 54 |
| chr5_-_176057518 | 0.01 |
ENST00000393693.2
|
SNCB
|
synuclein, beta |
| chr8_-_103668114 | 0.01 |
ENST00000285407.6
|
KLF10
|
Kruppel-like factor 10 |
| chr19_-_4043154 | 0.01 |
ENST00000535853.1
|
AC016586.1
|
AC016586.1 |
| chr4_+_128982430 | 0.01 |
ENST00000512292.1
ENST00000508819.1 |
LARP1B
|
La ribonucleoprotein domain family, member 1B |
| chr20_+_44462481 | 0.01 |
ENST00000491381.1
ENST00000342644.5 ENST00000372542.1 |
SNX21
|
sorting nexin family member 21 |
| chr4_-_152329987 | 0.01 |
ENST00000508847.1
|
RP11-610P16.1
|
RP11-610P16.1 |
| chr14_-_77608121 | 0.01 |
ENST00000319374.4
|
ZDHHC22
|
zinc finger, DHHC-type containing 22 |
| chr10_+_135340859 | 0.01 |
ENST00000252945.3
ENST00000421586.1 ENST00000418356.1 |
CYP2E1
|
cytochrome P450, family 2, subfamily E, polypeptide 1 |
| chr9_+_131549610 | 0.01 |
ENST00000223865.8
|
TBC1D13
|
TBC1 domain family, member 13 |
| chr3_+_196439275 | 0.01 |
ENST00000296333.5
|
PIGX
|
phosphatidylinositol glycan anchor biosynthesis, class X |
| chr11_-_6495082 | 0.01 |
ENST00000536344.1
|
TRIM3
|
tripartite motif containing 3 |
| chr7_-_23053693 | 0.01 |
ENST00000409763.1
ENST00000409923.1 |
FAM126A
|
family with sequence similarity 126, member A |
| chr7_-_30029328 | 0.01 |
ENST00000425819.2
ENST00000409570.1 |
SCRN1
|
secernin 1 |
| chr7_-_23053719 | 0.01 |
ENST00000432176.2
ENST00000440481.1 |
FAM126A
|
family with sequence similarity 126, member A |
| chrX_+_49028265 | 0.01 |
ENST00000376322.3
ENST00000376327.5 |
PLP2
|
proteolipid protein 2 (colonic epithelium-enriched) |
| chr9_+_112542591 | 0.01 |
ENST00000483909.1
ENST00000314527.4 ENST00000413420.1 ENST00000302798.7 ENST00000555236.1 ENST00000510514.5 |
PALM2
PALM2-AKAP2
AKAP2
|
paralemmin 2 PALM2-AKAP2 readthrough A kinase (PRKA) anchor protein 2 |
| chr12_-_125348329 | 0.01 |
ENST00000546215.1
ENST00000415380.2 ENST00000261693.6 ENST00000376788.1 ENST00000545493.1 |
SCARB1
|
scavenger receptor class B, member 1 |
| chr4_+_152330390 | 0.01 |
ENST00000503146.1
ENST00000435205.1 |
FAM160A1
|
family with sequence similarity 160, member A1 |
| chr19_+_10531150 | 0.01 |
ENST00000352831.6
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr8_+_145065705 | 0.01 |
ENST00000533044.1
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr15_-_59225844 | 0.01 |
ENST00000380516.2
|
SLTM
|
SAFB-like, transcription modulator |
| chr2_-_46769694 | 0.01 |
ENST00000522587.1
|
ATP6V1E2
|
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 |
| chr8_+_22423168 | 0.01 |
ENST00000518912.1
ENST00000428103.1 |
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr3_-_114173530 | 0.00 |
ENST00000470311.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr16_+_3014217 | 0.00 |
ENST00000572045.1
|
KREMEN2
|
kringle containing transmembrane protein 2 |
| chr11_-_64510409 | 0.00 |
ENST00000394429.1
ENST00000394428.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr7_-_27219849 | 0.00 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr11_-_107729504 | 0.00 |
ENST00000265836.7
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr15_-_66649010 | 0.00 |
ENST00000367709.4
ENST00000261881.4 |
TIPIN
|
TIMELESS interacting protein |
| chr6_-_35480705 | 0.00 |
ENST00000229771.6
|
TULP1
|
tubby like protein 1 |
| chr11_-_1036706 | 0.00 |
ENST00000421673.2
|
MUC6
|
mucin 6, oligomeric mucus/gel-forming |
| chr3_-_52931557 | 0.00 |
ENST00000504329.1
ENST00000355083.5 |
TMEM110-MUSTN1
TMEM110
|
TMEM110-MUSTN1 readthrough transmembrane protein 110 |
| chr19_+_14544099 | 0.00 |
ENST00000242783.6
ENST00000586557.1 ENST00000590097.1 |
PKN1
|
protein kinase N1 |
| chr5_+_169011033 | 0.00 |
ENST00000513795.1
|
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr8_+_145065521 | 0.00 |
ENST00000534791.1
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr2_+_62932779 | 0.00 |
ENST00000427809.1
ENST00000405482.1 ENST00000431489.1 |
EHBP1
|
EH domain binding protein 1 |
| chr10_+_17271266 | 0.00 |
ENST00000224237.5
|
VIM
|
vimentin |
| chr19_+_41770236 | 0.00 |
ENST00000392006.3
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr19_+_20011775 | 0.00 |
ENST00000592245.1
ENST00000592160.1 ENST00000343769.5 |
AC007204.2
ZNF93
|
AC007204.2 zinc finger protein 93 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF423
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.3 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.0 | 0.1 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.1 | GO:0033383 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.0 | 0.0 | GO:0038183 | bile acid signaling pathway(GO:0038183) |
| 0.0 | 0.0 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.1 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.1 | GO:0033597 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0052726 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.0 | 0.1 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.0 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.0 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |