Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
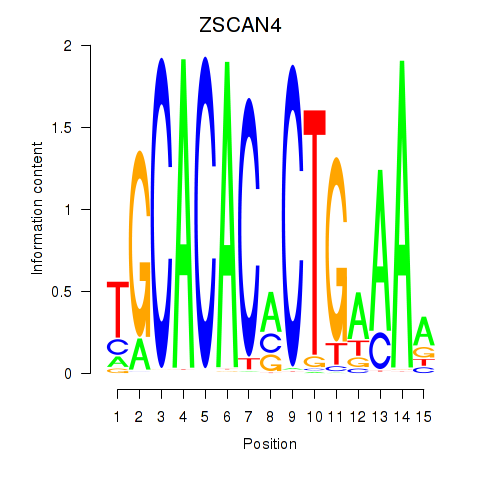
Results for ZSCAN4
Z-value: 0.62
Transcription factors associated with ZSCAN4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZSCAN4
|
ENSG00000180532.6 | zinc finger and SCAN domain containing 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZSCAN4 | hg19_v2_chr19_+_58180303_58180303 | 0.19 | 7.2e-01 | Click! |
Activity profile of ZSCAN4 motif
Sorted Z-values of ZSCAN4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_75062730 | 0.68 |
ENST00000420843.2
ENST00000360025.3 |
ARRB1
|
arrestin, beta 1 |
| chrX_-_20134990 | 0.58 |
ENST00000379651.3
ENST00000443379.3 ENST00000379643.5 |
MAP7D2
|
MAP7 domain containing 2 |
| chrX_+_3189861 | 0.52 |
ENST00000457435.1
ENST00000420429.2 |
CXorf28
|
chromosome X open reading frame 28 |
| chr3_+_111393659 | 0.52 |
ENST00000477665.1
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chrX_-_20134713 | 0.44 |
ENST00000452324.3
|
MAP7D2
|
MAP7 domain containing 2 |
| chr17_-_19648683 | 0.36 |
ENST00000573368.1
ENST00000457500.2 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr17_-_19648916 | 0.36 |
ENST00000444455.1
ENST00000439102.2 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr1_+_104615595 | 0.35 |
ENST00000418362.1
|
RP11-364B6.1
|
RP11-364B6.1 |
| chr2_+_240323439 | 0.34 |
ENST00000428471.1
ENST00000413029.1 |
AC062017.1
|
Uncharacterized protein |
| chr17_-_19648416 | 0.32 |
ENST00000426645.2
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr16_+_16481306 | 0.30 |
ENST00000422673.2
|
NPIPA7
|
nuclear pore complex interacting protein family, member A7 |
| chr2_+_189156389 | 0.29 |
ENST00000409843.1
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr9_-_93727673 | 0.24 |
ENST00000427745.1
|
RP11-367F23.1
|
RP11-367F23.1 |
| chr19_-_40596828 | 0.24 |
ENST00000414720.2
ENST00000455521.1 ENST00000340963.5 ENST00000595773.1 |
ZNF780A
|
zinc finger protein 780A |
| chr19_-_40596767 | 0.24 |
ENST00000599972.1
ENST00000450241.2 ENST00000595687.2 |
ZNF780A
|
zinc finger protein 780A |
| chr12_+_54694979 | 0.23 |
ENST00000552848.1
|
COPZ1
|
coatomer protein complex, subunit zeta 1 |
| chr14_+_71679350 | 0.22 |
ENST00000553621.1
|
RP6-91H8.1
|
RP6-91H8.1 |
| chr11_-_128812744 | 0.21 |
ENST00000458238.2
ENST00000531399.1 ENST00000602346.1 |
TP53AIP1
|
tumor protein p53 regulated apoptosis inducing protein 1 |
| chr9_+_34652164 | 0.21 |
ENST00000441545.2
ENST00000553620.1 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr20_+_23168759 | 0.20 |
ENST00000411595.1
|
RP4-737E23.2
|
RP4-737E23.2 |
| chr6_-_153304697 | 0.20 |
ENST00000367241.3
|
FBXO5
|
F-box protein 5 |
| chr17_+_30348024 | 0.20 |
ENST00000327564.7
ENST00000584368.1 ENST00000394713.3 ENST00000341671.7 |
LRRC37B
|
leucine rich repeat containing 37B |
| chr3_+_9944492 | 0.19 |
ENST00000383814.3
ENST00000454190.2 ENST00000454992.1 |
IL17RE
|
interleukin 17 receptor E |
| chr11_+_111750206 | 0.19 |
ENST00000530214.1
ENST00000530799.1 |
C11orf1
|
chromosome 11 open reading frame 1 |
| chr3_-_180397256 | 0.19 |
ENST00000442201.2
|
CCDC39
|
coiled-coil domain containing 39 |
| chr14_-_55658252 | 0.17 |
ENST00000395425.2
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr1_+_89829610 | 0.17 |
ENST00000370456.4
ENST00000535065.1 |
GBP6
|
guanylate binding protein family, member 6 |
| chr2_-_98972468 | 0.17 |
ENST00000454230.1
|
AC092675.3
|
Uncharacterized protein |
| chr1_-_8000872 | 0.17 |
ENST00000377507.3
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr1_-_228603694 | 0.16 |
ENST00000366697.2
|
TRIM17
|
tripartite motif containing 17 |
| chr21_+_25801041 | 0.16 |
ENST00000453784.2
ENST00000423581.1 |
AP000476.1
|
AP000476.1 |
| chr19_-_40562063 | 0.16 |
ENST00000598845.1
ENST00000593605.1 ENST00000221355.6 ENST00000434248.1 |
ZNF780B
|
zinc finger protein 780B |
| chr3_+_9944303 | 0.16 |
ENST00000421412.1
ENST00000295980.3 |
IL17RE
|
interleukin 17 receptor E |
| chr9_+_128510454 | 0.15 |
ENST00000491787.3
ENST00000447726.2 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr12_-_10022735 | 0.15 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr17_-_75878647 | 0.15 |
ENST00000374983.2
|
FLJ45079
|
FLJ45079 |
| chr1_+_94883991 | 0.14 |
ENST00000370214.4
|
ABCD3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr19_+_44764031 | 0.13 |
ENST00000592581.1
ENST00000590668.1 ENST00000588489.1 ENST00000391958.2 |
ZNF233
|
zinc finger protein 233 |
| chr14_+_89290965 | 0.13 |
ENST00000345383.5
ENST00000536576.1 ENST00000346301.4 ENST00000338104.6 ENST00000354441.6 ENST00000380656.2 ENST00000556651.1 ENST00000554686.1 |
TTC8
|
tetratricopeptide repeat domain 8 |
| chr1_-_169337176 | 0.13 |
ENST00000472647.1
ENST00000367811.3 |
NME7
|
NME/NM23 family member 7 |
| chr12_+_41086215 | 0.12 |
ENST00000547702.1
ENST00000551424.1 |
CNTN1
|
contactin 1 |
| chr17_-_46799872 | 0.12 |
ENST00000290294.3
|
PRAC1
|
prostate cancer susceptibility candidate 1 |
| chr5_+_61602055 | 0.12 |
ENST00000381103.2
|
KIF2A
|
kinesin heavy chain member 2A |
| chrX_+_1387693 | 0.12 |
ENST00000381529.3
ENST00000432318.2 ENST00000361536.3 ENST00000501036.2 ENST00000381524.3 ENST00000412290.1 |
CSF2RA
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr11_-_33774944 | 0.12 |
ENST00000532057.1
ENST00000531080.1 |
FBXO3
|
F-box protein 3 |
| chr1_+_94883931 | 0.11 |
ENST00000394233.2
ENST00000454898.2 ENST00000536817.1 |
ABCD3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr1_-_21978312 | 0.11 |
ENST00000359708.4
ENST00000290101.4 |
RAP1GAP
|
RAP1 GTPase activating protein |
| chr19_+_14491948 | 0.11 |
ENST00000358600.3
|
CD97
|
CD97 molecule |
| chr15_+_63413990 | 0.11 |
ENST00000261893.4
|
LACTB
|
lactamase, beta |
| chr11_+_61560348 | 0.11 |
ENST00000535723.1
ENST00000574708.1 |
FEN1
FADS2
|
flap structure-specific endonuclease 1 fatty acid desaturase 2 |
| chr13_-_99404875 | 0.10 |
ENST00000376503.5
|
SLC15A1
|
solute carrier family 15 (oligopeptide transporter), member 1 |
| chr21_-_33651324 | 0.10 |
ENST00000290130.3
|
MIS18A
|
MIS18 kinetochore protein A |
| chr21_+_45879814 | 0.10 |
ENST00000596691.1
|
LRRC3DN
|
LRRC3 downstream neighbor (non-protein coding) |
| chr19_-_35626104 | 0.10 |
ENST00000310123.3
ENST00000392225.3 |
LGI4
|
leucine-rich repeat LGI family, member 4 |
| chr2_-_175462934 | 0.10 |
ENST00000392546.2
ENST00000436221.1 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr17_+_63133587 | 0.10 |
ENST00000449996.3
ENST00000262406.9 |
RGS9
|
regulator of G-protein signaling 9 |
| chr21_-_46221684 | 0.10 |
ENST00000330942.5
|
UBE2G2
|
ubiquitin-conjugating enzyme E2G 2 |
| chr4_-_89619386 | 0.10 |
ENST00000323061.5
|
NAP1L5
|
nucleosome assembly protein 1-like 5 |
| chr2_-_70520539 | 0.09 |
ENST00000482975.2
ENST00000438261.1 |
SNRPG
|
small nuclear ribonucleoprotein polypeptide G |
| chr12_-_96390108 | 0.09 |
ENST00000538703.1
ENST00000261208.3 |
HAL
|
histidine ammonia-lyase |
| chr20_+_43343517 | 0.09 |
ENST00000372865.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr15_+_63414017 | 0.09 |
ENST00000413507.2
|
LACTB
|
lactamase, beta |
| chr19_+_44716678 | 0.09 |
ENST00000586228.1
ENST00000588219.1 ENST00000313040.7 ENST00000589707.1 ENST00000588394.1 ENST00000589005.1 |
ZNF227
|
zinc finger protein 227 |
| chr4_-_168155577 | 0.09 |
ENST00000541354.1
ENST00000509854.1 ENST00000512681.1 ENST00000421836.2 ENST00000510741.1 ENST00000510403.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr4_-_103682071 | 0.09 |
ENST00000505239.1
|
MANBA
|
mannosidase, beta A, lysosomal |
| chr10_-_62332357 | 0.09 |
ENST00000503366.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr13_+_60971427 | 0.09 |
ENST00000535286.1
ENST00000377881.2 |
TDRD3
|
tudor domain containing 3 |
| chr4_-_103682145 | 0.09 |
ENST00000226578.4
|
MANBA
|
mannosidase, beta A, lysosomal |
| chr13_+_60971080 | 0.08 |
ENST00000377894.2
|
TDRD3
|
tudor domain containing 3 |
| chr19_+_44716768 | 0.08 |
ENST00000586048.1
|
ZNF227
|
zinc finger protein 227 |
| chr15_+_32885657 | 0.08 |
ENST00000448387.2
ENST00000569659.1 |
GOLGA8N
|
golgin A8 family, member N |
| chr4_-_186578674 | 0.08 |
ENST00000438278.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr11_+_61717842 | 0.08 |
ENST00000449131.2
|
BEST1
|
bestrophin 1 |
| chr2_-_224810070 | 0.08 |
ENST00000429915.1
ENST00000233055.4 |
WDFY1
|
WD repeat and FYVE domain containing 1 |
| chr9_+_128509624 | 0.08 |
ENST00000342287.5
ENST00000373487.4 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr19_+_17516624 | 0.08 |
ENST00000596322.1
ENST00000600008.1 ENST00000601885.1 |
CTD-2521M24.9
|
CTD-2521M24.9 |
| chr8_+_107670064 | 0.07 |
ENST00000312046.6
|
OXR1
|
oxidation resistance 1 |
| chr17_+_73089382 | 0.07 |
ENST00000538213.2
ENST00000584118.1 |
SLC16A5
|
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr13_+_114238997 | 0.07 |
ENST00000538138.1
ENST00000375370.5 |
TFDP1
|
transcription factor Dp-1 |
| chr20_-_16554078 | 0.07 |
ENST00000354981.2
ENST00000355755.3 ENST00000378003.2 ENST00000408042.1 |
KIF16B
|
kinesin family member 16B |
| chr4_+_74718906 | 0.07 |
ENST00000226524.3
|
PF4V1
|
platelet factor 4 variant 1 |
| chr16_+_89228757 | 0.07 |
ENST00000565008.1
|
LINC00304
|
long intergenic non-protein coding RNA 304 |
| chr17_-_34207295 | 0.07 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr1_+_52195542 | 0.07 |
ENST00000462759.1
ENST00000486942.1 |
OSBPL9
|
oxysterol binding protein-like 9 |
| chr4_-_168155730 | 0.07 |
ENST00000502330.1
ENST00000357154.3 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr6_+_90272027 | 0.07 |
ENST00000522441.1
|
ANKRD6
|
ankyrin repeat domain 6 |
| chr10_-_120514720 | 0.07 |
ENST00000369151.3
ENST00000340214.4 |
CACUL1
|
CDK2-associated, cullin domain 1 |
| chr1_+_231376941 | 0.07 |
ENST00000436239.1
ENST00000366647.4 ENST00000366646.3 ENST00000416000.1 |
GNPAT
|
glyceronephosphate O-acyltransferase |
| chr15_+_76135622 | 0.07 |
ENST00000338677.4
ENST00000267938.4 ENST00000569423.1 |
UBE2Q2
|
ubiquitin-conjugating enzyme E2Q family member 2 |
| chr18_+_32173276 | 0.07 |
ENST00000591816.1
ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA
|
dystrobrevin, alpha |
| chr12_-_96390063 | 0.07 |
ENST00000541929.1
|
HAL
|
histidine ammonia-lyase |
| chr12_-_11548496 | 0.07 |
ENST00000389362.4
ENST00000565533.1 ENST00000546254.1 |
PRB2
PRB1
|
proline-rich protein BstNI subfamily 2 proline-rich protein BstNI subfamily 1 |
| chr7_-_135194822 | 0.06 |
ENST00000428680.2
ENST00000315544.5 ENST00000423368.2 ENST00000451834.1 ENST00000361528.4 ENST00000356162.4 ENST00000541284.1 |
CNOT4
|
CCR4-NOT transcription complex, subunit 4 |
| chr9_+_125137565 | 0.06 |
ENST00000373698.5
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chrX_+_56259316 | 0.06 |
ENST00000468660.1
|
KLF8
|
Kruppel-like factor 8 |
| chr8_+_26247878 | 0.06 |
ENST00000518611.1
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr19_-_38714847 | 0.05 |
ENST00000420980.2
ENST00000355526.4 |
DPF1
|
D4, zinc and double PHD fingers family 1 |
| chr2_+_237994519 | 0.05 |
ENST00000392008.2
ENST00000409334.1 ENST00000409629.1 |
COPS8
|
COP9 signalosome subunit 8 |
| chr15_-_82641706 | 0.05 |
ENST00000439287.4
|
GOLGA6L10
|
golgin A6 family-like 10 |
| chr11_+_61717535 | 0.05 |
ENST00000534553.1
ENST00000301774.9 |
BEST1
|
bestrophin 1 |
| chr11_+_111807863 | 0.05 |
ENST00000440460.2
|
DIXDC1
|
DIX domain containing 1 |
| chr12_+_16064106 | 0.05 |
ENST00000428559.2
|
DERA
|
deoxyribose-phosphate aldolase (putative) |
| chr10_-_30348439 | 0.05 |
ENST00000375377.1
|
KIAA1462
|
KIAA1462 |
| chr2_-_160473114 | 0.05 |
ENST00000392783.2
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr7_-_10979750 | 0.05 |
ENST00000339600.5
|
NDUFA4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4, 9kDa |
| chr10_+_80828774 | 0.05 |
ENST00000334512.5
|
ZMIZ1
|
zinc finger, MIZ-type containing 1 |
| chr13_+_28813645 | 0.05 |
ENST00000282391.5
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr10_-_101989315 | 0.05 |
ENST00000370397.7
|
CHUK
|
conserved helix-loop-helix ubiquitous kinase |
| chr4_+_15779901 | 0.05 |
ENST00000226279.3
|
CD38
|
CD38 molecule |
| chr4_-_168155169 | 0.05 |
ENST00000534949.1
ENST00000535728.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr12_+_133613878 | 0.04 |
ENST00000392319.2
ENST00000543758.1 |
ZNF84
|
zinc finger protein 84 |
| chr2_-_101034070 | 0.04 |
ENST00000264249.3
|
CHST10
|
carbohydrate sulfotransferase 10 |
| chr15_+_75550940 | 0.04 |
ENST00000300576.5
|
GOLGA6C
|
golgin A6 family, member C |
| chr12_+_16064258 | 0.04 |
ENST00000524480.1
ENST00000531803.1 ENST00000532964.1 |
DERA
|
deoxyribose-phosphate aldolase (putative) |
| chr17_-_73761222 | 0.04 |
ENST00000437911.1
ENST00000225614.2 |
GALK1
|
galactokinase 1 |
| chr4_-_168155300 | 0.04 |
ENST00000541637.1
|
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr9_-_20621834 | 0.04 |
ENST00000429426.2
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr12_-_57634475 | 0.04 |
ENST00000393825.1
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 |
| chr6_+_114178512 | 0.04 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr15_-_37392724 | 0.04 |
ENST00000424352.2
|
MEIS2
|
Meis homeobox 2 |
| chr5_+_149887672 | 0.03 |
ENST00000261797.6
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr2_-_167232484 | 0.03 |
ENST00000375387.4
ENST00000303354.6 ENST00000409672.1 |
SCN9A
|
sodium channel, voltage-gated, type IX, alpha subunit |
| chr2_-_214016314 | 0.03 |
ENST00000434687.1
ENST00000374319.4 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr6_+_90272339 | 0.03 |
ENST00000522779.1
|
ANKRD6
|
ankyrin repeat domain 6 |
| chr11_-_77791156 | 0.03 |
ENST00000281031.4
|
NDUFC2
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2, 14.5kDa |
| chr7_-_99381884 | 0.03 |
ENST00000336411.2
|
CYP3A4
|
cytochrome P450, family 3, subfamily A, polypeptide 4 |
| chr2_+_183580954 | 0.03 |
ENST00000264065.7
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10 |
| chr14_+_53173890 | 0.03 |
ENST00000445930.2
|
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr3_-_121740969 | 0.03 |
ENST00000393631.1
ENST00000273691.3 ENST00000344209.5 |
ILDR1
|
immunoglobulin-like domain containing receptor 1 |
| chr19_-_11266471 | 0.03 |
ENST00000592540.1
|
SPC24
|
SPC24, NDC80 kinetochore complex component |
| chr22_+_18121562 | 0.03 |
ENST00000355028.3
|
BCL2L13
|
BCL2-like 13 (apoptosis facilitator) |
| chr12_-_48226897 | 0.03 |
ENST00000434070.1
|
HDAC7
|
histone deacetylase 7 |
| chr15_-_37392703 | 0.02 |
ENST00000382766.2
ENST00000444725.1 |
MEIS2
|
Meis homeobox 2 |
| chr2_+_234545092 | 0.02 |
ENST00000344644.5
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr1_+_233765353 | 0.02 |
ENST00000366620.1
|
KCNK1
|
potassium channel, subfamily K, member 1 |
| chrX_+_17393543 | 0.02 |
ENST00000380060.3
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies) |
| chr19_-_3025614 | 0.02 |
ENST00000447365.2
|
TLE2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) |
| chr19_+_7660716 | 0.02 |
ENST00000160298.4
ENST00000446248.2 |
CAMSAP3
|
calmodulin regulated spectrin-associated protein family, member 3 |
| chr12_-_86230315 | 0.02 |
ENST00000361228.3
|
RASSF9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| chr1_+_2036149 | 0.02 |
ENST00000482686.1
ENST00000400920.1 ENST00000486681.1 |
PRKCZ
|
protein kinase C, zeta |
| chr16_+_16472912 | 0.02 |
ENST00000530217.2
|
NPIPA7
|
nuclear pore complex interacting protein family, member A7 |
| chr1_-_184006611 | 0.02 |
ENST00000546159.1
|
COLGALT2
|
collagen beta(1-O)galactosyltransferase 2 |
| chr5_+_31193847 | 0.02 |
ENST00000514738.1
ENST00000265071.2 |
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney) |
| chr3_+_130569592 | 0.02 |
ENST00000533801.2
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr8_-_16050142 | 0.02 |
ENST00000536385.1
ENST00000381998.4 |
MSR1
|
macrophage scavenger receptor 1 |
| chr16_-_86542455 | 0.02 |
ENST00000595886.1
ENST00000597578.1 ENST00000593604.1 |
FENDRR
|
FOXF1 adjacent non-coding developmental regulatory RNA |
| chr7_-_84569561 | 0.02 |
ENST00000439105.1
|
AC074183.4
|
AC074183.4 |
| chr20_-_60573188 | 0.02 |
ENST00000474089.1
|
TAF4
|
TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 135kDa |
| chr4_-_168155700 | 0.01 |
ENST00000357545.4
ENST00000512648.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr19_-_9649253 | 0.01 |
ENST00000593003.1
|
ZNF426
|
zinc finger protein 426 |
| chr16_+_23847267 | 0.01 |
ENST00000321728.7
|
PRKCB
|
protein kinase C, beta |
| chr15_-_82939157 | 0.01 |
ENST00000559949.1
|
RP13-996F3.5
|
golgin A6 family-like 18 |
| chr3_+_130569429 | 0.01 |
ENST00000505330.1
ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr9_-_107361788 | 0.01 |
ENST00000374779.2
|
OR13C5
|
olfactory receptor, family 13, subfamily C, member 5 |
| chr1_+_32645645 | 0.01 |
ENST00000373609.1
|
TXLNA
|
taxilin alpha |
| chr12_-_54694807 | 0.01 |
ENST00000435572.2
|
NFE2
|
nuclear factor, erythroid 2 |
| chr12_+_122242597 | 0.01 |
ENST00000267197.5
|
SETD1B
|
SET domain containing 1B |
| chr1_-_153599426 | 0.01 |
ENST00000392622.1
|
S100A13
|
S100 calcium binding protein A13 |
| chr3_+_52009110 | 0.01 |
ENST00000491470.1
|
ABHD14A
|
abhydrolase domain containing 14A |
| chr14_+_53173910 | 0.01 |
ENST00000606149.1
ENST00000555339.1 ENST00000556813.1 |
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr2_+_95831529 | 0.01 |
ENST00000295210.6
ENST00000453539.2 |
ZNF2
|
zinc finger protein 2 |
| chr8_+_72755796 | 0.01 |
ENST00000524152.1
|
RP11-383H13.1
|
Protein LOC100132891; cDNA FLJ53548 |
| chr6_-_33548006 | 0.01 |
ENST00000374467.3
|
BAK1
|
BCL2-antagonist/killer 1 |
| chrX_+_119495934 | 0.01 |
ENST00000218008.3
ENST00000361319.3 ENST00000539306.1 |
ATP1B4
|
ATPase, Na+/K+ transporting, beta 4 polypeptide |
| chrX_-_133792480 | 0.01 |
ENST00000359237.4
|
PLAC1
|
placenta-specific 1 |
| chr10_-_103880209 | 0.01 |
ENST00000425280.1
|
LDB1
|
LIM domain binding 1 |
| chr11_-_62341445 | 0.01 |
ENST00000329251.4
|
EEF1G
|
eukaryotic translation elongation factor 1 gamma |
| chr21_-_44035168 | 0.01 |
ENST00000419628.1
|
AP001626.1
|
AP001626.1 |
| chr8_-_72987810 | 0.01 |
ENST00000262209.4
|
TRPA1
|
transient receptor potential cation channel, subfamily A, member 1 |
| chr7_-_30008849 | 0.00 |
ENST00000409497.1
|
SCRN1
|
secernin 1 |
| chr1_+_169337412 | 0.00 |
ENST00000426663.1
|
BLZF1
|
basic leucine zipper nuclear factor 1 |
| chr17_-_7080227 | 0.00 |
ENST00000574330.1
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr17_-_56405407 | 0.00 |
ENST00000343736.4
|
BZRAP1
|
benzodiazepine receptor (peripheral) associated protein 1 |
| chr1_+_159141397 | 0.00 |
ENST00000368124.4
ENST00000368125.4 ENST00000416746.1 |
CADM3
|
cell adhesion molecule 3 |
| chr11_+_61717816 | 0.00 |
ENST00000435278.2
|
BEST1
|
bestrophin 1 |
| chr19_+_49927006 | 0.00 |
ENST00000576655.1
|
CTD-3148I10.1
|
golgi-associated, olfactory signaling regulator |
| chr2_-_239198743 | 0.00 |
ENST00000440245.1
ENST00000431832.1 |
PER2
|
period circadian clock 2 |
| chr6_+_130742231 | 0.00 |
ENST00000545622.1
|
TMEM200A
|
transmembrane protein 200A |
| chr22_+_18121356 | 0.00 |
ENST00000317582.5
ENST00000543133.1 ENST00000538149.1 ENST00000337612.5 ENST00000493680.1 |
BCL2L13
|
BCL2-like 13 (apoptosis facilitator) |
| chr20_+_51588873 | 0.00 |
ENST00000371497.5
|
TSHZ2
|
teashirt zinc finger homeobox 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZSCAN4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.1 | 0.2 | GO:0007387 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.1 | 0.2 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.1 | 0.2 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.3 | GO:0097398 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.2 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.1 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.0 | 0.1 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.2 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.2 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.0 | 0.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.1 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.0 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.1 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.2 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.0 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0031893 | vasopressin receptor binding(GO:0031893) V2 vasopressin receptor binding(GO:0031896) |
| 0.1 | 1.0 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 0.2 | GO:0004397 | histidine ammonia-lyase activity(GO:0004397) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.3 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0005427 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.0 | 0.1 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.0 | 0.2 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) linoleoyl-CoA desaturase activity(GO:0016213) 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.0 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.0 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.0 | 0.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |