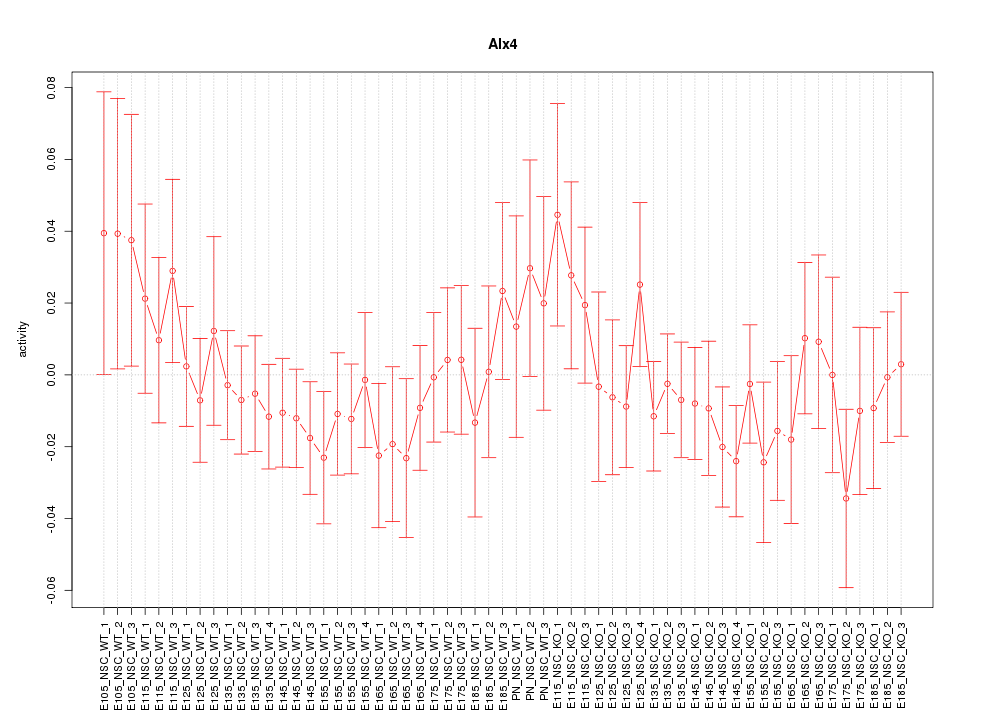
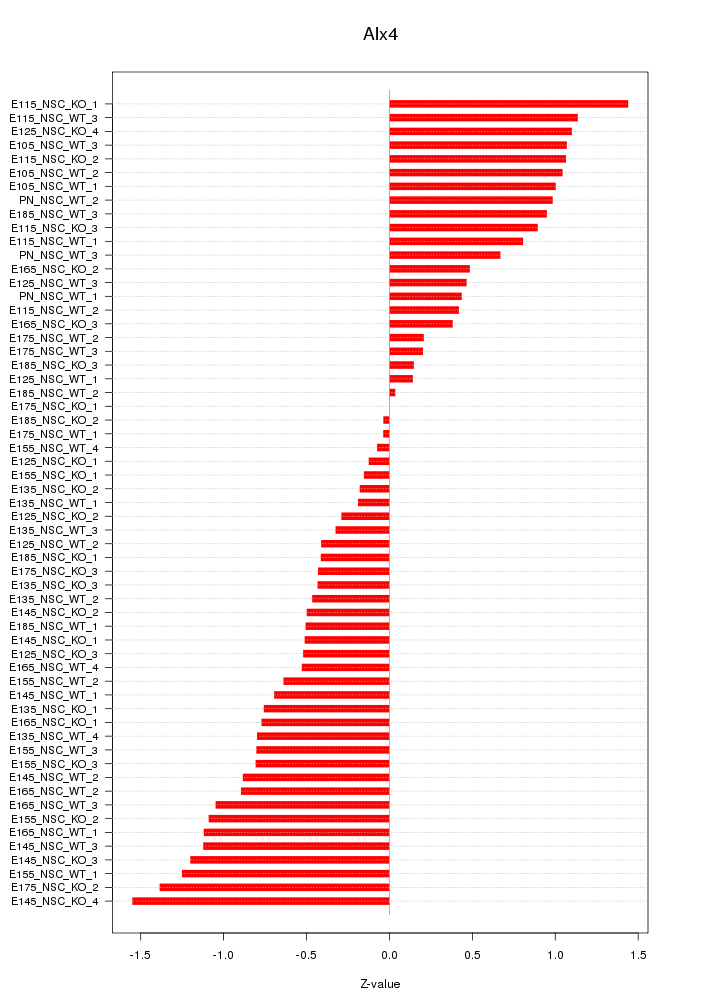
Motif ID: Alx4
Z-value: 0.760

Transcription factors associated with Alx4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Alx4 | ENSMUSG00000040310.6 | Alx4 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Alx4 | mm10_v2_chr2_+_93642307_93642388 | 0.48 | 9.9e-05 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.2 | GO:0072204 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 1.4 | 4.3 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 1.4 | 4.1 | GO:0061743 | motor learning(GO:0061743) |
| 1.3 | 2.6 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 1.1 | 4.4 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) |
| 0.9 | 3.5 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.8 | 2.5 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.8 | 2.4 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.8 | 4.7 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.7 | 2.2 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.7 | 6.7 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.7 | 2.9 | GO:0019287 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.7 | 2.9 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.7 | 3.5 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.7 | 2.0 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) olefin metabolic process(GO:1900673) |
| 0.6 | 1.9 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.6 | 7.9 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.6 | 1.8 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.6 | 2.2 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.5 | 4.2 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.5 | 1.9 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.4 | 1.2 | GO:0050929 | corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.4 | 2.7 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.4 | 1.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.3 | 3.0 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.3 | 1.7 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.3 | 2.0 | GO:0036233 | glycine import(GO:0036233) |
| 0.3 | 2.2 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.3 | 0.9 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.3 | 2.2 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.3 | 2.0 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.3 | 2.5 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.3 | 4.9 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.2 | 0.7 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.2 | 3.6 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.2 | 0.9 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.2 | 1.6 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.2 | 1.8 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.2 | 1.8 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.2 | 0.9 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.2 | 1.7 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.2 | 1.3 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.2 | 3.7 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.2 | 4.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.2 | 2.7 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.2 | 3.6 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.2 | 1.8 | GO:0046036 | GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.2 | 0.9 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.2 | 0.7 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.2 | 3.6 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.2 | 1.9 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.2 | 1.0 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.2 | 4.0 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.1 | 0.6 | GO:0018158 | protein oxidation(GO:0018158) |
| 0.1 | 2.9 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.1 | 0.4 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.8 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.1 | 0.5 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.1 | 0.4 | GO:0010749 | regulation of nitric oxide mediated signal transduction(GO:0010749) |
| 0.1 | 0.6 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.1 | 0.5 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.1 | 2.1 | GO:0006221 | pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.1 | 1.6 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.1 | 0.6 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 0.3 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 4.4 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 3.2 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.1 | 1.5 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 1.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.4 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 0.9 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.1 | 0.6 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.1 | 0.9 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.1 | 0.7 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.1 | 1.1 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.1 | 0.7 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.1 | 1.6 | GO:1903427 | negative regulation of reactive oxygen species biosynthetic process(GO:1903427) |
| 0.1 | 0.3 | GO:0006547 | histidine metabolic process(GO:0006547) |
| 0.1 | 3.6 | GO:0042491 | auditory receptor cell differentiation(GO:0042491) |
| 0.1 | 1.2 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.1 | 2.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 0.8 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 1.1 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 1.1 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.1 | 3.8 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.1 | 1.3 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.1 | 2.2 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 0.6 | GO:0071774 | response to fibroblast growth factor(GO:0071774) |
| 0.1 | 0.7 | GO:0021549 | cerebellum development(GO:0021549) metencephalon development(GO:0022037) |
| 0.1 | 0.3 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.1 | 0.7 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 1.8 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 0.7 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 1.0 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.0 | 1.3 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 1.0 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.4 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 2.7 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.3 | GO:0072539 | T-helper 17 cell differentiation(GO:0072539) |
| 0.0 | 0.3 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 1.0 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 1.6 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 1.0 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 2.1 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.0 | 1.9 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 2.3 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 0.9 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.9 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 1.1 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.3 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.3 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.2 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.1 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.2 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.1 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.0 | 1.6 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 1.8 | GO:0001942 | hair follicle development(GO:0001942) molting cycle process(GO:0022404) hair cycle process(GO:0022405) skin epidermis development(GO:0098773) |
| 0.0 | 2.7 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 0.1 | GO:0060768 | epithelial cell proliferation involved in prostate gland development(GO:0060767) regulation of epithelial cell proliferation involved in prostate gland development(GO:0060768) negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 1.0 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.6 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 2.8 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.3 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.0 | 1.0 | GO:0045580 | regulation of T cell differentiation(GO:0045580) |
| 0.0 | 1.3 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.2 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.0 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.3 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 6.7 | GO:0006412 | translation(GO:0006412) |
| 0.0 | 0.4 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.0 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.0 | 0.7 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.4 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.9 | 3.6 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.9 | 3.4 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 0.8 | 4.1 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.7 | 4.0 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.7 | 2.0 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.5 | 7.9 | GO:0042555 | MCM complex(GO:0042555) |
| 0.5 | 4.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.3 | 1.8 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.3 | 2.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.3 | 1.6 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.2 | 0.7 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.2 | 1.0 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.2 | 1.7 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.2 | 2.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 2.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.2 | 1.0 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.2 | 4.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.2 | 3.3 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.2 | 1.0 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.2 | 1.6 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.6 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 0.9 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.7 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 1.0 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 1.7 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 1.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 1.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.9 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 4.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 2.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.1 | 0.3 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 4.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 1.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 1.0 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.1 | 2.8 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 1.6 | GO:0045120 | pronucleus(GO:0045120) |
| 0.1 | 1.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 1.0 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 2.7 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 0.6 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 1.7 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 1.1 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 3.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 0.5 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 2.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 1.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 13.0 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 8.5 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 3.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.5 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 3.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 2.5 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 1.9 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 1.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.5 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 2.4 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.6 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 1.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.2 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 2.1 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 2.5 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 5.4 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.7 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.4 | GO:0005113 | patched binding(GO:0005113) |
| 0.8 | 2.4 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.6 | 1.8 | GO:0052740 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.6 | 1.8 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.6 | 3.5 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.6 | 4.0 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.5 | 1.6 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.5 | 6.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.5 | 2.0 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.4 | 2.7 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.3 | 1.4 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.3 | 2.0 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.3 | 2.0 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.3 | 2.9 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.3 | 0.9 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.3 | 2.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.3 | 3.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.3 | 2.5 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.2 | 3.6 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.2 | 1.9 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.2 | 1.8 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.2 | 3.5 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.2 | 1.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.2 | 1.6 | GO:0018741 | alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 0.2 | 1.6 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.2 | 2.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.2 | 0.6 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.2 | 4.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.2 | 1.0 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.2 | 3.4 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.2 | 1.7 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.2 | 0.7 | GO:0051378 | primary amine oxidase activity(GO:0008131) serotonin binding(GO:0051378) |
| 0.2 | 1.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.2 | 7.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.2 | 4.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.2 | 0.7 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.2 | 1.8 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 0.9 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.2 | 0.6 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 7.9 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 6.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.9 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.4 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.1 | 3.0 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.4 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.1 | 0.8 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.1 | 4.3 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 4.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 1.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.7 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 2.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 2.2 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 2.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 4.7 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.1 | 1.8 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 2.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 1.0 | GO:0034951 | pivalyl-CoA mutase activity(GO:0034784) o-hydroxylaminobenzoate mutase activity(GO:0034951) lupeol synthase activity(GO:0042299) beta-amyrin synthase activity(GO:0042300) baruol synthase activity(GO:0080011) |
| 0.1 | 0.8 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.3 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.2 | GO:0044388 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) small protein activating enzyme binding(GO:0044388) cupric ion binding(GO:1903135) cuprous ion binding(GO:1903136) glyoxalase (glycolic acid-forming) activity(GO:1990422) |
| 0.1 | 3.9 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 0.5 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 0.7 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 2.6 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 2.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.7 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 1.6 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) |
| 0.1 | 0.3 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.1 | 1.0 | GO:0052890 | oxidoreductase activity, acting on the CH-CH group of donors, with a flavin as acceptor(GO:0052890) |
| 0.1 | 1.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 2.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.0 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 2.7 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 2.0 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.3 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.0 | 1.6 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.9 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 4.5 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 6.9 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 11.8 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.2 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) |
| 0.0 | 0.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 3.6 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 1.5 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 2.1 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 2.2 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 1.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 6.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.3 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.0 | 0.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.6 | GO:0043774 | UDP-N-acetylmuramoylalanyl-D-glutamyl-2,6-diaminopimelate-D-alanyl-D-alanine ligase activity(GO:0008766) ribosomal S6-glutamic acid ligase activity(GO:0018169) coenzyme F420-0 gamma-glutamyl ligase activity(GO:0043773) coenzyme F420-2 alpha-glutamyl ligase activity(GO:0043774) protein-glycine ligase activity(GO:0070735) protein-glycine ligase activity, initiating(GO:0070736) protein-glycine ligase activity, elongating(GO:0070737) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 0.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |