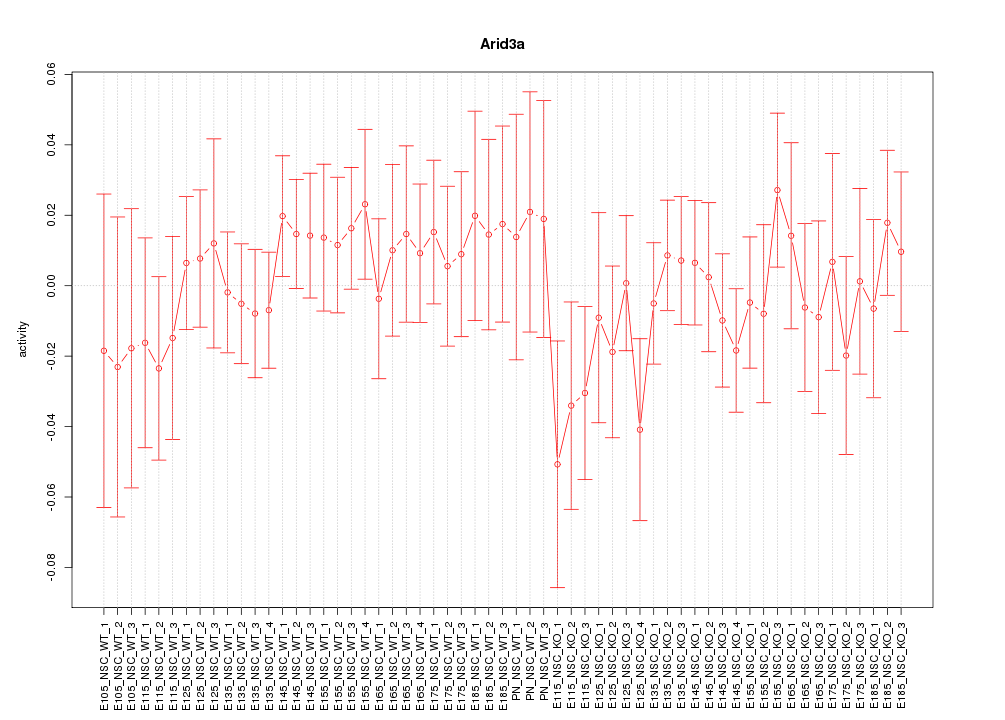
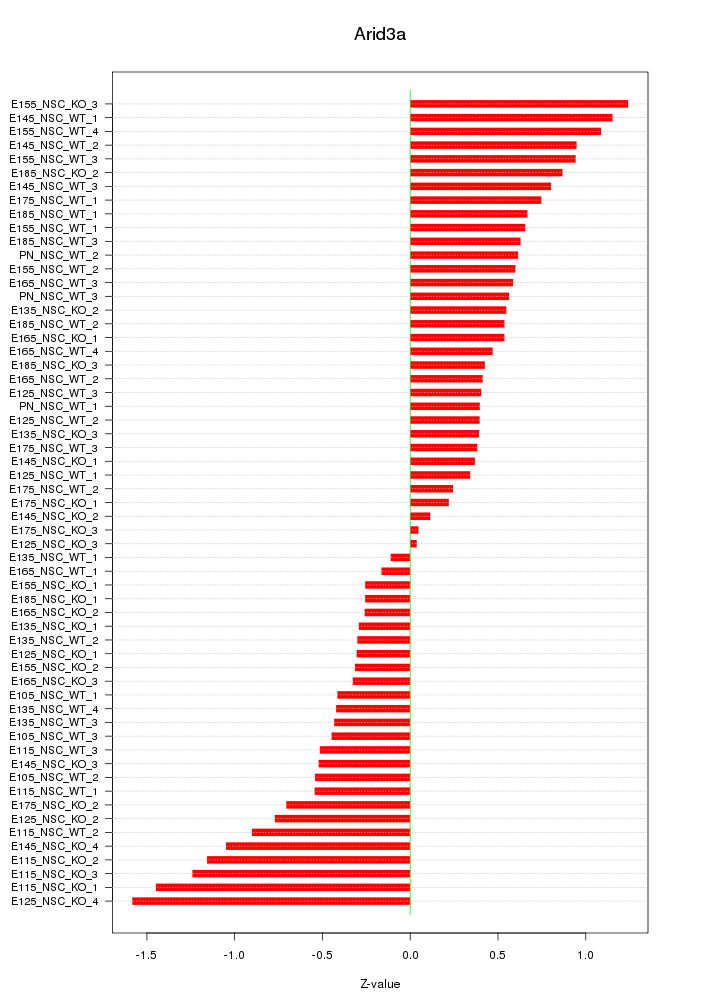
Motif ID: Arid3a
Z-value: 0.666
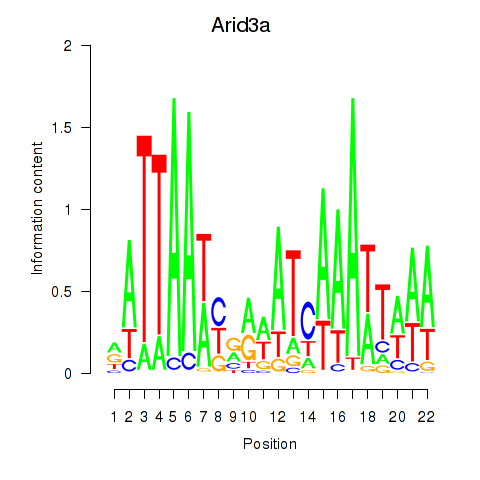
Transcription factors associated with Arid3a:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Arid3a | ENSMUSG00000019564.6 | Arid3a |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Arid3a | mm10_v2_chr10_+_79927039_79927139 | -0.61 | 3.8e-07 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.3 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.6 | 1.7 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.4 | 1.6 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.4 | 1.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.3 | 2.6 | GO:0007379 | segment specification(GO:0007379) |
| 0.2 | 2.3 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.2 | 0.8 | GO:0072592 | regulation of integrin biosynthetic process(GO:0045113) oxygen metabolic process(GO:0072592) |
| 0.2 | 0.6 | GO:0032240 | RNA import into nucleus(GO:0006404) mRNA export from nucleus in response to heat stress(GO:0031990) negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.2 | 0.7 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 0.6 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 3.1 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.1 | 1.5 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.1 | 3.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.9 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.1 | 0.4 | GO:0032382 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) positive regulation of cholesterol import(GO:1904109) positive regulation of sterol import(GO:2000911) |
| 0.1 | 2.7 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.9 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.4 | GO:0042167 | heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 1.0 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.3 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 1.6 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.4 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.4 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.6 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 1.8 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.0 | 0.7 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 1.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.8 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 2.2 | GO:0046323 | glucose import(GO:0046323) |
| 0.0 | 1.1 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.3 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.4 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 3.2 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.4 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.2 | 4.1 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.2 | 0.8 | GO:0071438 | NADPH oxidase complex(GO:0043020) invadopodium membrane(GO:0071438) |
| 0.1 | 0.6 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 1.6 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 2.7 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.7 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 0.4 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 1.5 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.9 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 0.4 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.1 | 2.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 1.6 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.9 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 1.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 3.1 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.8 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 2.2 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0004096 | catalase activity(GO:0004096) |
| 0.4 | 1.5 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.3 | 0.9 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.3 | 0.8 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.2 | 0.7 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) |
| 0.2 | 4.1 | GO:0050811 | GABA-A receptor activity(GO:0004890) GABA receptor binding(GO:0050811) |
| 0.2 | 1.7 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.2 | 2.2 | GO:0005536 | glucose binding(GO:0005536) |
| 0.1 | 0.6 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.4 | GO:0033814 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.1 | 1.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.4 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.1 | 2.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 2.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 1.0 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.1 | 1.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 4.0 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 1.7 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 2.5 | GO:0008757 | S-adenosylmethionine-dependent methyltransferase activity(GO:0008757) |
| 0.0 | 2.4 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 3.3 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |