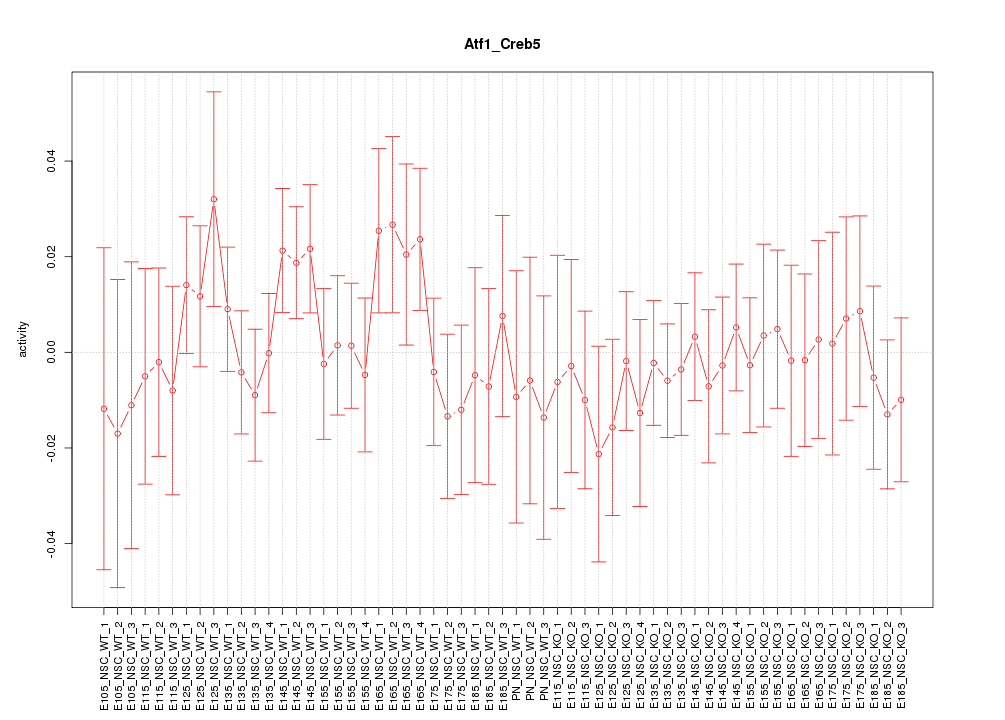
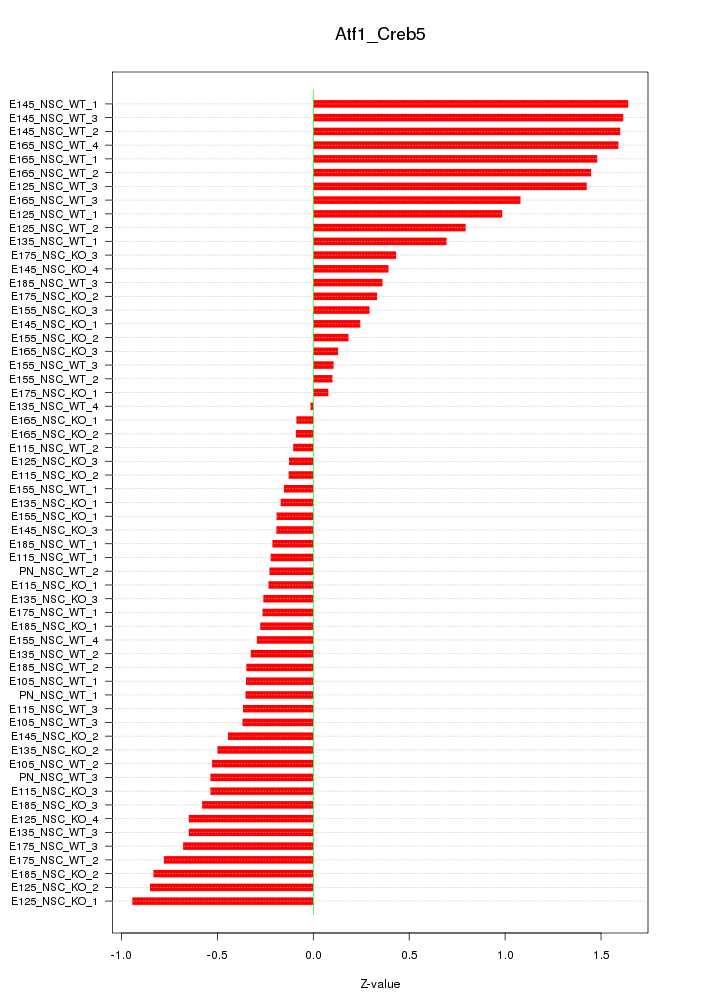
Motif ID: Atf1_Creb5
Z-value: 0.690
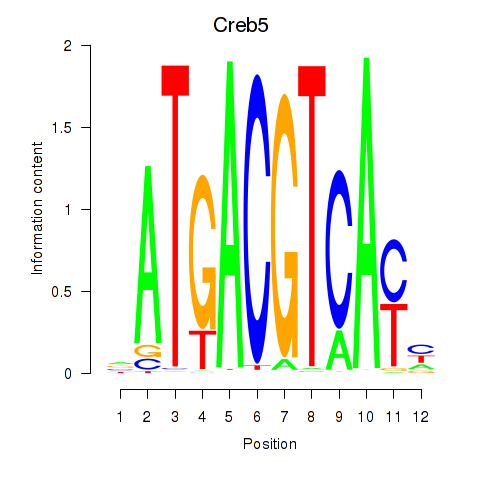
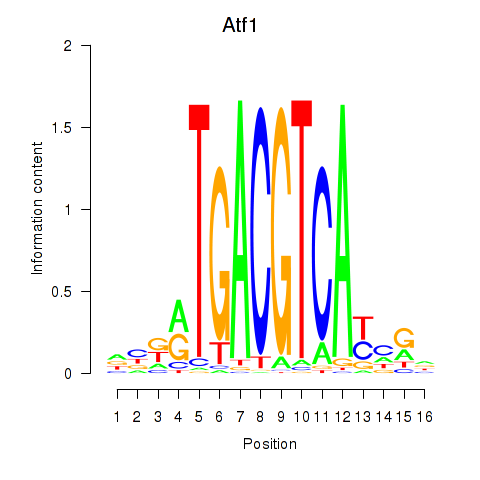
Transcription factors associated with Atf1_Creb5:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Atf1 | ENSMUSG00000023027.6 | Atf1 |
| Creb5 | ENSMUSG00000053007.6 | Creb5 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Atf1 | mm10_v2_chr15_+_100227871_100227878 | -0.24 | 6.7e-02 | Click! |
| Creb5 | mm10_v2_chr6_+_53573364_53573394 | -0.13 | 3.2e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 1.2 | 3.6 | GO:0010958 | regulation of amino acid import(GO:0010958) regulation of L-arginine import(GO:0010963) |
| 1.2 | 3.5 | GO:1900736 | regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 0.7 | 2.7 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.5 | 1.6 | GO:0031038 | myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.5 | 1.5 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.5 | 4.3 | GO:0060373 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.5 | 2.3 | GO:0051012 | microtubule sliding(GO:0051012) negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.5 | 5.0 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.4 | 4.4 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.4 | 1.2 | GO:0043379 | memory T cell differentiation(GO:0043379) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.4 | 1.2 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.4 | 7.4 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.4 | 2.2 | GO:0035743 | CD4-positive, alpha-beta T cell cytokine production(GO:0035743) T-helper 2 cell cytokine production(GO:0035745) |
| 0.3 | 2.9 | GO:0030432 | peristalsis(GO:0030432) |
| 0.3 | 1.5 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.3 | 0.9 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.3 | 0.9 | GO:0060022 | hard palate development(GO:0060022) soft palate development(GO:0060023) |
| 0.3 | 0.8 | GO:0009182 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) GDP metabolic process(GO:0046710) |
| 0.3 | 0.8 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.3 | 1.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.3 | 2.1 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.3 | 3.0 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.2 | 3.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.2 | 1.0 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.2 | 4.5 | GO:0001553 | luteinization(GO:0001553) |
| 0.2 | 0.7 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.2 | 4.6 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.2 | 1.4 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.2 | 0.9 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.2 | 1.5 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.2 | 1.0 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.2 | 0.8 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.2 | 0.6 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.2 | 0.8 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.2 | 0.9 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.2 | 0.7 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.2 | 1.0 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.2 | 0.5 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.2 | 1.4 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.2 | 0.7 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.2 | 0.8 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.2 | 0.5 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.2 | 0.5 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.2 | 0.5 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.1 | 1.4 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 1.9 | GO:0030825 | positive regulation of cGMP metabolic process(GO:0030825) positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.1 | 0.9 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.1 | 1.3 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.1 | 0.5 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 0.7 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 | 1.5 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.3 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 | 1.1 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 1.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.4 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.6 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 1.0 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 1.5 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.1 | 0.3 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.1 | 0.5 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.1 | 0.5 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.1 | 0.6 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.1 | 0.9 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.1 | 0.3 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.1 | 0.6 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.1 | 0.3 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 | 0.3 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.1 | 0.7 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.2 | GO:0021764 | amygdala development(GO:0021764) |
| 0.1 | 0.2 | GO:1900060 | negative regulation of sphingolipid biosynthetic process(GO:0090155) cellular sphingolipid homeostasis(GO:0090156) negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.1 | 0.6 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 2.4 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.6 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 0.6 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 0.3 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.1 | 0.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.7 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 0.2 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.1 | 0.4 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.1 | 0.5 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 0.6 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 0.3 | GO:1900116 | sequestering of extracellular ligand from receptor(GO:0035581) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.3 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 0.4 | GO:0051231 | spindle elongation(GO:0051231) regulation of oocyte maturation(GO:1900193) |
| 0.1 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.1 | 0.7 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 3.7 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.1 | 0.9 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.8 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 5.3 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.3 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 0.2 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.8 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.2 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.3 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.9 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.3 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 1.4 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.5 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 3.2 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.1 | GO:0046077 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.0 | 1.1 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 2.2 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.2 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 1.7 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.3 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.8 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.2 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 1.0 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.0 | 0.3 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 1.4 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.5 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 0.6 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.3 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 1.3 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.8 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 1.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.8 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.6 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.1 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.0 | 0.3 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.0 | 0.3 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 1.8 | GO:0046888 | negative regulation of hormone secretion(GO:0046888) |
| 0.0 | 0.5 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.3 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 2.8 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.4 | GO:0030813 | positive regulation of nucleotide catabolic process(GO:0030813) positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.0 | 0.1 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 | 0.4 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.7 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.5 | GO:0006862 | nucleotide transport(GO:0006862) |
| 0.0 | 1.0 | GO:1902850 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 | 0.5 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.3 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.5 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.0 | 0.8 | GO:0051591 | response to cAMP(GO:0051591) |
| 0.0 | 0.6 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 1.4 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.0 | 0.4 | GO:0006754 | ATP biosynthetic process(GO:0006754) |
| 0.0 | 0.1 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.5 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.0 | 0.1 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.9 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 | 0.5 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.5 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.0 | 0.0 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 2.1 | GO:0006275 | regulation of DNA replication(GO:0006275) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 1.0 | GO:0071774 | response to fibroblast growth factor(GO:0071774) |
| 0.0 | 0.7 | GO:0000423 | macromitophagy(GO:0000423) mitophagy in response to mitochondrial depolarization(GO:0098779) response to mitochondrial depolarisation(GO:0098780) |
| 0.0 | 0.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.0 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.2 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.3 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 | 1.2 | GO:0015758 | glucose transport(GO:0015758) |
| 0.0 | 0.6 | GO:0035904 | aorta development(GO:0035904) |
| 0.0 | 0.1 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.4 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.0 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.5 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.3 | 5.1 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.3 | 3.5 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.3 | 5.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.3 | 2.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.2 | 0.9 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 2.4 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.2 | 0.9 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.2 | 2.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 0.6 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.2 | 0.6 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.2 | 2.7 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 2.0 | GO:0016589 | NURF complex(GO:0016589) |
| 0.2 | 3.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.7 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.5 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.1 | 1.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.7 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 1.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 0.8 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 0.7 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 7.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 0.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 1.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.6 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.9 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 0.6 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 1.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 0.5 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 1.0 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.1 | 3.2 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.2 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.1 | 2.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 4.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 0.3 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 0.6 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.3 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.2 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.8 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.4 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.0 | 2.2 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 1.1 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.5 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.6 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 2.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 2.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.4 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 1.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.8 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.6 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.1 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 4.0 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.3 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 1.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.5 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.7 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 2.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.6 | 1.9 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.6 | 4.3 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.6 | 3.6 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.6 | 1.7 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.5 | 4.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.5 | 1.6 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.5 | 1.5 | GO:0033699 | DNA 5'-adenosine monophosphate hydrolase activity(GO:0033699) |
| 0.4 | 3.5 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.3 | 6.0 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.2 | 2.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.2 | 1.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 0.9 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.2 | 0.9 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.2 | 4.9 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.2 | 0.6 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.2 | 0.8 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.2 | 0.6 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.2 | 0.6 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 4.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 0.9 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.2 | 0.5 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.2 | 2.0 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.7 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 1.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 2.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.7 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 1.2 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.1 | 0.5 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.1 | 1.6 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.1 | 0.3 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 0.3 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 0.9 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.2 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.1 | 1.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.4 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 5.1 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 0.4 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.4 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.1 | 0.5 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 2.3 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 0.9 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.5 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 3.6 | GO:0018602 | sulfonate dioxygenase activity(GO:0000907) 2,4-dichlorophenoxyacetate alpha-ketoglutarate dioxygenase activity(GO:0018602) hypophosphite dioxygenase activity(GO:0034792) gibberellin 2-beta-dioxygenase activity(GO:0045543) C-19 gibberellin 2-beta-dioxygenase activity(GO:0052634) C-20 gibberellin 2-beta-dioxygenase activity(GO:0052635) |
| 0.1 | 0.4 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.7 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 2.7 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.7 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 3.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 0.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.7 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.3 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.1 | 4.2 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 1.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.9 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 1.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 2.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.6 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 0.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.5 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.6 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.3 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 0.8 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 5.3 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.1 | 0.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.5 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 4.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.8 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 8.3 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 1.1 | GO:0034930 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) heparan sulfate 2-O-sulfotransferase activity(GO:0004394) HNK-1 sulfotransferase activity(GO:0016232) heparan sulfate 6-O-sulfotransferase activity(GO:0017095) trans-9R,10R-dihydrodiolphenanthrene sulfotransferase activity(GO:0018721) 1-phenanthrol sulfotransferase activity(GO:0018722) 3-phenanthrol sulfotransferase activity(GO:0018723) 4-phenanthrol sulfotransferase activity(GO:0018724) trans-3,4-dihydrodiolphenanthrene sulfotransferase activity(GO:0018725) 9-phenanthrol sulfotransferase activity(GO:0018726) 2-phenanthrol sulfotransferase activity(GO:0018727) phenanthrol sulfotransferase activity(GO:0019111) 1-hydroxypyrene sulfotransferase activity(GO:0034930) proteoglycan sulfotransferase activity(GO:0050698) cholesterol sulfotransferase activity(GO:0051922) hydroxyjasmonate sulfotransferase activity(GO:0080131) |
| 0.0 | 0.8 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.2 | GO:0004120 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.0 | 0.3 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.0 | 1.2 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.0 | 0.5 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.4 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.3 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.8 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 1.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.7 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 1.8 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 1.0 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.3 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0070492 | sialic acid binding(GO:0033691) oligosaccharide binding(GO:0070492) |
| 0.0 | 2.2 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 4.6 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.7 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.1 | GO:0003905 | alkylbase DNA N-glycosylase activity(GO:0003905) DNA-3-methylbase glycosylase activity(GO:0043733) |
| 0.0 | 0.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.6 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0034943 | acyl-CoA ligase activity(GO:0003996) 3-oxo-2-(2'-pentenyl)cyclopentane-1-octanoic acid CoA ligase activity(GO:0010435) 3-isopropenyl-6-oxoheptanoyl-CoA synthetase activity(GO:0018854) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA synthetase activity(GO:0018855) benzoyl acetate-CoA ligase activity(GO:0018856) 2,4-dichlorobenzoate-CoA ligase activity(GO:0018857) pivalate-CoA ligase activity(GO:0034783) cyclopropanecarboxylate-CoA ligase activity(GO:0034793) adipate-CoA ligase activity(GO:0034796) citronellyl-CoA ligase activity(GO:0034823) mentha-1,3-dione-CoA ligase activity(GO:0034841) thiophene-2-carboxylate-CoA ligase activity(GO:0034842) 2,4,4-trimethylpentanoate-CoA ligase activity(GO:0034865) cis-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034942) trans-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034943) branched-chain acyl-CoA synthetase (ADP-forming) activity(GO:0043759) aryl-CoA synthetase (ADP-forming) activity(GO:0043762) 3-hydroxypropionyl-CoA synthetase activity(GO:0043955) perillic acid:CoA ligase (ADP-forming) activity(GO:0052685) perillic acid:CoA ligase (AMP-forming) activity(GO:0052686) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (ADP-forming) activity(GO:0052687) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (AMP-forming) activity(GO:0052688) pristanate-CoA ligase activity(GO:0070251) malonyl-CoA synthetase activity(GO:0090409) |
| 0.0 | 0.8 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |