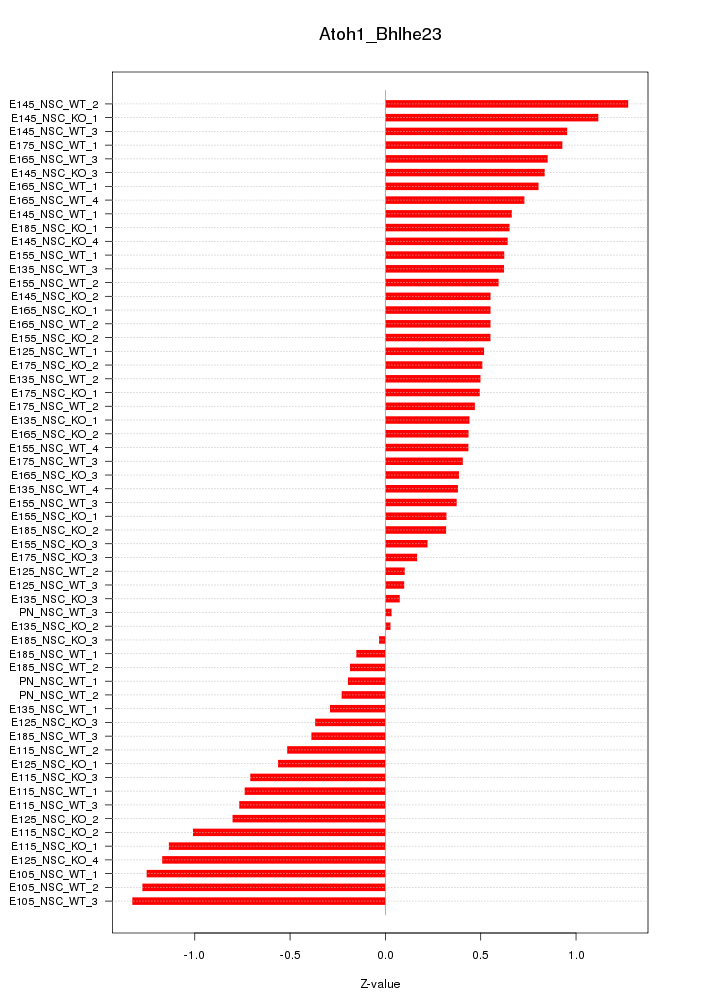
Motif ID: Atoh1_Bhlhe23
Z-value: 0.659
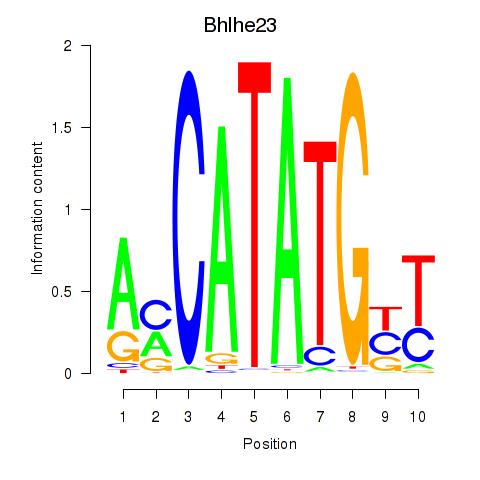
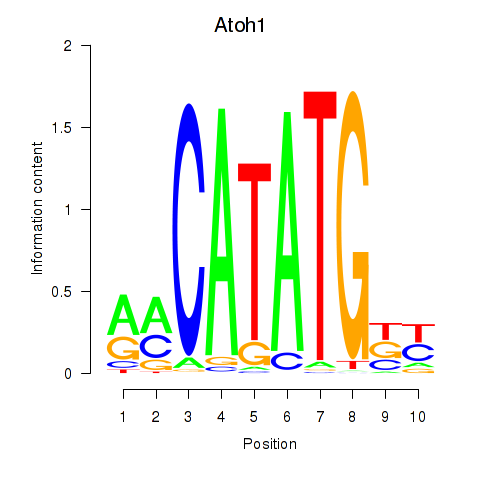
Transcription factors associated with Atoh1_Bhlhe23:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Atoh1 | ENSMUSG00000073043.4 | Atoh1 |
| Bhlhe23 | ENSMUSG00000045493.3 | Bhlhe23 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bhlhe23 | mm10_v2_chr2_-_180776900_180776900 | -0.15 | 2.6e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.6 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 2.1 | 8.4 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 2.0 | 8.0 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 1.5 | 7.3 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 1.1 | 3.2 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.6 | 2.5 | GO:1903288 | regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.6 | 7.5 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.5 | 8.7 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.4 | 2.5 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.4 | 2.5 | GO:0032796 | uropod organization(GO:0032796) |
| 0.4 | 1.2 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.4 | 3.2 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.3 | 2.6 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.3 | 1.4 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.3 | 3.1 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.3 | 2.8 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.2 | 0.7 | GO:0032240 | RNA import into nucleus(GO:0006404) mRNA export from nucleus in response to heat stress(GO:0031990) negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.2 | 1.2 | GO:0042701 | progesterone secretion(GO:0042701) |
| 0.2 | 2.5 | GO:0030836 | positive regulation of actin filament depolymerization(GO:0030836) |
| 0.2 | 18.1 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.2 | 1.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.2 | 0.9 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.2 | 1.5 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.2 | 1.1 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.2 | 1.9 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 2.0 | GO:0072010 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 | 3.2 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 1.8 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.1 | 1.0 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 2.9 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.1 | 2.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.8 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.1 | GO:1900020 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) hypophysis morphogenesis(GO:0048850) cervix development(GO:0060067) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.6 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.9 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 5.7 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 2.9 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.0 | 2.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.8 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 0.3 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.6 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 1.8 | GO:0010508 | autophagosome assembly(GO:0000045) positive regulation of autophagy(GO:0010508) |
| 0.0 | 1.7 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 4.9 | GO:0043523 | regulation of neuron apoptotic process(GO:0043523) |
| 0.0 | 0.7 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.1 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.6 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 4.4 | GO:0007409 | axonogenesis(GO:0007409) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 8.7 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.4 | 7.3 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.4 | 2.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.4 | 18.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.3 | 5.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 2.5 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.2 | 2.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 7.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 2.5 | GO:0070938 | contractile ring(GO:0070938) |
| 0.2 | 6.1 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.2 | 3.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 3.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.7 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 2.4 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 2.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 1.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 1.1 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 2.9 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 0.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 2.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.7 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 6.6 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 3.2 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 2.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 1.8 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.3 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 1.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 2.2 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.9 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 1.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.7 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.3 | 5.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.3 | 2.0 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.3 | 7.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.3 | 2.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.3 | 14.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.3 | 2.5 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.2 | 2.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 1.1 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.2 | 2.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 0.6 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.2 | 4.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.7 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.9 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 4.9 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 5.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 2.9 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 2.9 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 1.4 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 3.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 8.6 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 0.6 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.7 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.9 | GO:0034237 | AMP binding(GO:0016208) protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.8 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 1.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 2.7 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 3.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.2 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 2.5 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 19.3 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 2.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 2.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.8 | GO:0044212 | transcription regulatory region DNA binding(GO:0044212) |