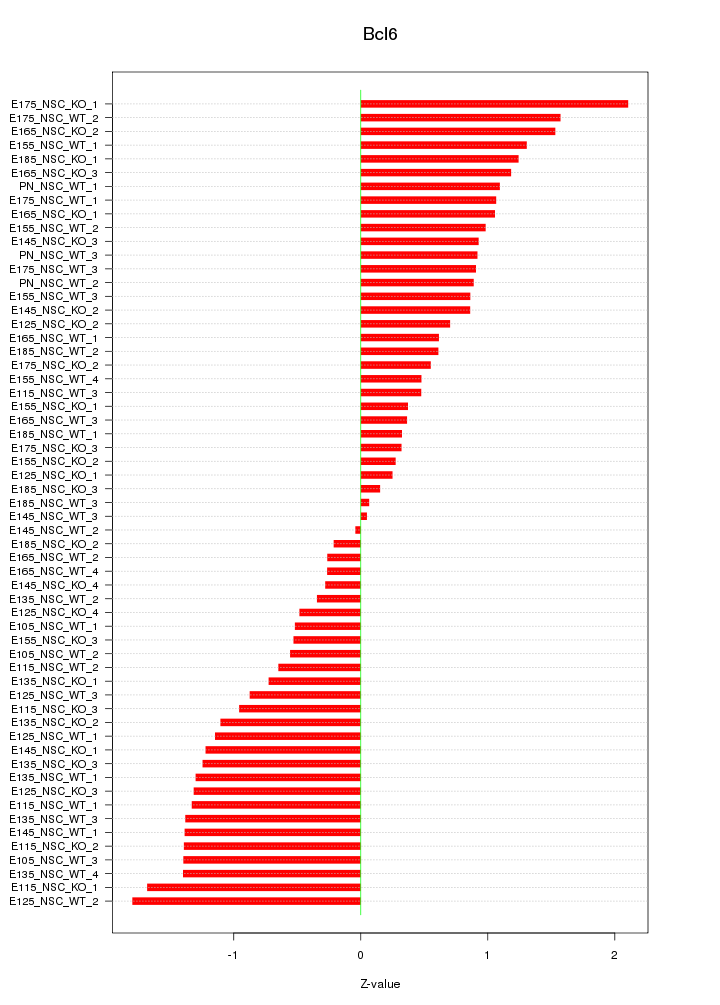
Motif ID: Bcl6
Z-value: 0.978
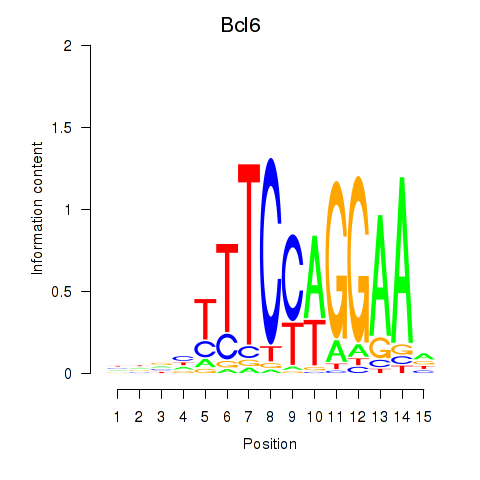
Transcription factors associated with Bcl6:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Bcl6 | ENSMUSG00000022508.5 | Bcl6 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bcl6 | mm10_v2_chr16_-_23988852_23988852 | 0.64 | 4.2e-08 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 12.5 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 2.0 | 7.9 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 1.7 | 6.8 | GO:0042938 | dipeptide transport(GO:0042938) |
| 1.6 | 11.3 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 1.4 | 4.3 | GO:0010751 | regulation of arginine metabolic process(GO:0000821) negative regulation of nitric oxide mediated signal transduction(GO:0010751) negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 1.3 | 3.8 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 1.2 | 13.3 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 1.0 | 4.0 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 1.0 | 4.9 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.9 | 2.7 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.9 | 3.4 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.9 | 4.3 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.8 | 3.1 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.7 | 4.4 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.7 | 2.1 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.7 | 5.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.6 | 6.0 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.6 | 8.0 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.6 | 1.7 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.5 | 1.1 | GO:2000587 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.5 | 2.5 | GO:0090164 | asymmetric Golgi ribbon formation(GO:0090164) |
| 0.5 | 2.0 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.4 | 2.5 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.4 | 2.5 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.4 | 1.2 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.4 | 0.8 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.4 | 4.2 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.4 | 1.9 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) positive regulation of receptor binding(GO:1900122) |
| 0.3 | 3.5 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.3 | 8.8 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.3 | 1.3 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.3 | 1.3 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.3 | 2.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.3 | 1.2 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.3 | 6.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.3 | 0.8 | GO:0070973 | COPI-coated vesicle budding(GO:0035964) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.3 | 2.7 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 4.7 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.2 | 3.4 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.2 | 1.6 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.2 | 10.6 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.2 | 1.5 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.2 | 0.6 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.2 | 1.6 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.2 | 3.9 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.2 | 1.0 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.2 | 1.3 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.2 | 1.5 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.2 | 1.2 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.1 | 1.3 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.1 | 0.5 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 4.6 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 0.7 | GO:0032026 | response to magnesium ion(GO:0032026) |
| 0.1 | 2.2 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.8 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.1 | 5.5 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 1.4 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 2.5 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.1 | 0.7 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.4 | GO:0036438 | maintenance of lens transparency(GO:0036438) membrane fission(GO:0090148) ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.1 | 3.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 1.0 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.1 | 0.7 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.1 | 1.3 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 0.8 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 0.9 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.1 | 2.8 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.1 | 0.2 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.1 | 2.3 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 7.2 | GO:0099643 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.1 | 0.2 | GO:0071726 | response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.1 | 4.0 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 0.5 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.3 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.6 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 2.0 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 0.5 | GO:1902287 | semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.1 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.0 | 0.7 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.8 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.7 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 1.6 | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway(GO:2001237) |
| 0.0 | 0.3 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.1 | GO:0034143 | regulation of toll-like receptor 4 signaling pathway(GO:0034143) |
| 0.0 | 0.3 | GO:1903749 | regulation of protein targeting to mitochondrion(GO:1903214) positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 1.9 | GO:0010506 | regulation of autophagy(GO:0010506) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.8 | 13.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.7 | 3.5 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.5 | 7.9 | GO:0043205 | fibril(GO:0043205) |
| 0.4 | 2.0 | GO:0001652 | granular component(GO:0001652) |
| 0.4 | 1.2 | GO:0044299 | C-fiber(GO:0044299) |
| 0.3 | 1.7 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.3 | 1.3 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.3 | 6.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.3 | 8.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 4.9 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.2 | 0.7 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.2 | 2.5 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.2 | 2.9 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.2 | 4.3 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.2 | 2.2 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.2 | 0.5 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.1 | 6.4 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 3.9 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 0.9 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 0.7 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.1 | 4.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 3.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 3.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.7 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 3.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 3.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 2.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.4 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 14.7 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 1.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 4.6 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.1 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 1.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 1.8 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 3.4 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 11.3 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 2.1 | 12.5 | GO:0045545 | syndecan binding(GO:0045545) |
| 1.9 | 13.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 1.6 | 7.9 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 1.4 | 6.8 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 1.1 | 8.0 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 1.0 | 6.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.9 | 3.4 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.8 | 3.9 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.8 | 5.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.7 | 4.9 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.7 | 4.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.7 | 4.6 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.6 | 2.5 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.6 | 2.5 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.5 | 1.5 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.5 | 4.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.4 | 4.1 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.4 | 2.0 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.4 | 1.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.3 | 2.0 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.3 | 4.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.3 | 2.9 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.3 | 1.2 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.3 | 1.4 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.3 | 3.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.3 | 0.5 | GO:0005110 | frizzled-2 binding(GO:0005110) chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.3 | 3.2 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.3 | 0.8 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.3 | 0.8 | GO:0015173 | hydrogen:amino acid symporter activity(GO:0005280) aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.3 | 4.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 1.0 | GO:0015183 | sulfur amino acid transmembrane transporter activity(GO:0000099) L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.2 | 10.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 3.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 2.8 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 0.7 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.2 | 4.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.2 | 5.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 2.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.2 | 2.6 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.1 | 4.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.8 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 2.5 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 4.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 5.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 2.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 1.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.8 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 6.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 1.6 | GO:0030546 | receptor activator activity(GO:0030546) protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 1.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 2.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 2.5 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 3.9 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.1 | 0.2 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 5.2 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.1 | 1.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.6 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 1.7 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.9 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.5 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.8 | GO:0051723 | protein methylesterase activity(GO:0051723) |
| 0.0 | 0.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 1.6 | GO:0052771 | coenzyme F390-A hydrolase activity(GO:0052770) coenzyme F390-G hydrolase activity(GO:0052771) |
| 0.0 | 1.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 1.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.5 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.7 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.8 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 3.0 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) lipase activator activity(GO:0060229) |
| 0.0 | 0.6 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 1.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) exopeptidase activity(GO:0008238) |
| 0.0 | 1.8 | GO:0030246 | carbohydrate binding(GO:0030246) |