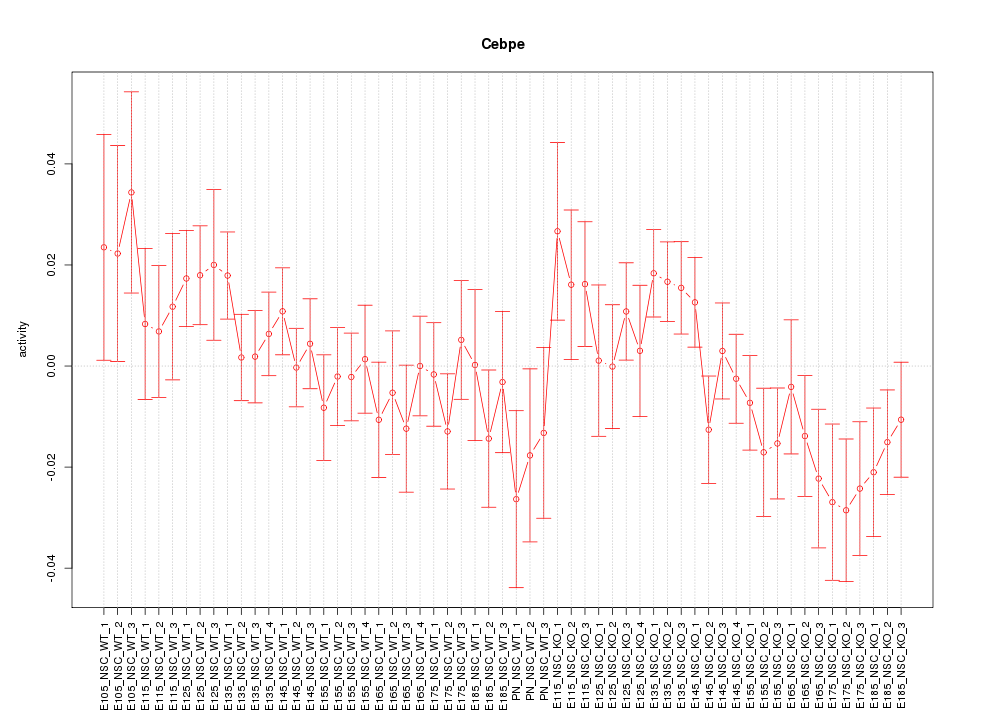
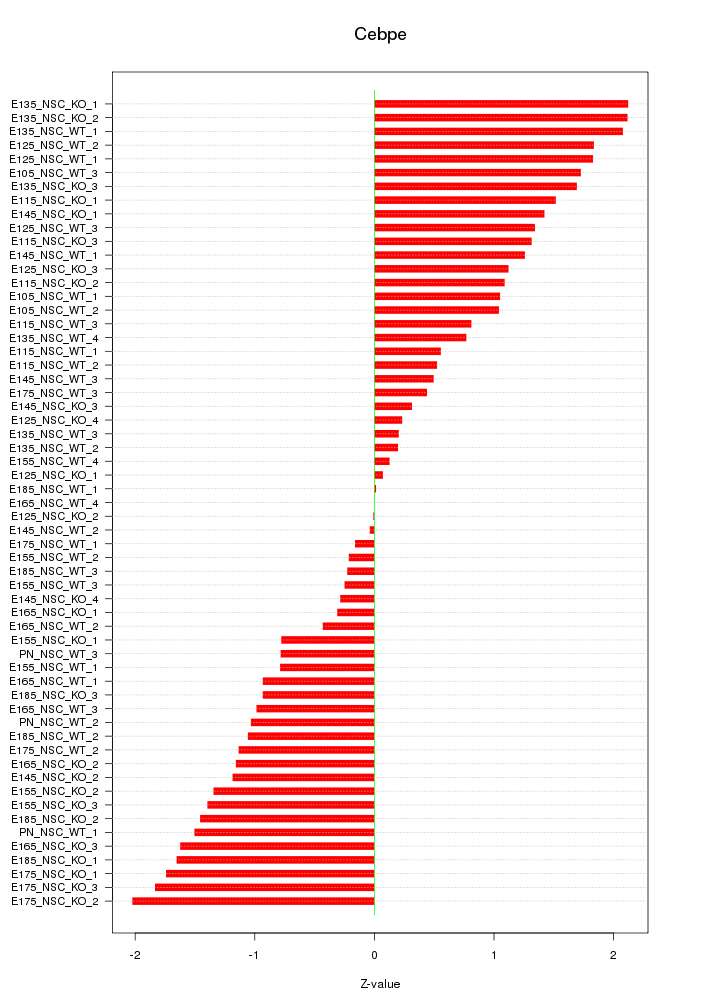
Motif ID: Cebpe
Z-value: 1.148

Transcription factors associated with Cebpe:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Cebpe | ENSMUSG00000052435.6 | Cebpe |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 17.7 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 4.6 | 23.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 2.2 | 6.6 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 2.2 | 8.6 | GO:1903898 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 1.6 | 4.8 | GO:0060166 | olfactory pit development(GO:0060166) |
| 1.4 | 1.4 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 1.4 | 4.1 | GO:0048818 | embryonic nail plate morphogenesis(GO:0035880) positive regulation of hair follicle maturation(GO:0048818) positive regulation of catagen(GO:0051795) frontal suture morphogenesis(GO:0060364) |
| 1.2 | 3.6 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 1.2 | 3.5 | GO:0097402 | neuroblast migration(GO:0097402) |
| 1.0 | 4.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 1.0 | 4.9 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 1.0 | 3.8 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.9 | 2.8 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.9 | 2.6 | GO:0030862 | neuroblast division in subventricular zone(GO:0021849) regulation of polarized epithelial cell differentiation(GO:0030860) positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.9 | 3.4 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.8 | 2.5 | GO:0009177 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) |
| 0.8 | 2.4 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.8 | 2.4 | GO:0002765 | immune response-inhibiting signal transduction(GO:0002765) |
| 0.8 | 8.1 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.8 | 3.1 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.8 | 1.6 | GO:0016102 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.8 | 7.0 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.8 | 3.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.8 | 3.8 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.8 | 2.3 | GO:0002295 | T-helper cell lineage commitment(GO:0002295) |
| 0.7 | 4.4 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.7 | 2.2 | GO:1901228 | positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.7 | 2.2 | GO:0030167 | proteoglycan catabolic process(GO:0030167) |
| 0.7 | 3.6 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.7 | 2.9 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.7 | 4.3 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.7 | 2.8 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.7 | 2.8 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.7 | 2.0 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.6 | 7.7 | GO:2000254 | regulation of male germ cell proliferation(GO:2000254) |
| 0.6 | 3.2 | GO:2000065 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.6 | 2.5 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.6 | 5.6 | GO:0085020 | protein K6-linked ubiquitination(GO:0085020) |
| 0.6 | 2.4 | GO:0032901 | positive regulation of neurotrophin production(GO:0032901) |
| 0.6 | 1.8 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.6 | 2.4 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.6 | 5.7 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.6 | 4.6 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.6 | 1.7 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.5 | 1.6 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.5 | 1.6 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.5 | 1.6 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.5 | 1.6 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.5 | 2.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.5 | 2.5 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) astrocyte activation(GO:0048143) regulation of resting membrane potential(GO:0060075) |
| 0.5 | 1.0 | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.5 | 2.5 | GO:0019659 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.5 | 3.4 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.5 | 2.9 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.5 | 1.0 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.5 | 1.8 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.5 | 1.8 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.4 | 2.6 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.4 | 5.9 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.4 | 1.7 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) resolution of recombination intermediates(GO:0071139) |
| 0.4 | 1.7 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.4 | 2.4 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.4 | 2.0 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.4 | 2.3 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.4 | 6.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.4 | 1.9 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.4 | 2.2 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.4 | 5.5 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.3 | 3.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.3 | 2.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.3 | 1.0 | GO:0030210 | heparin biosynthetic process(GO:0030210) Tie signaling pathway(GO:0048014) |
| 0.3 | 1.0 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.3 | 1.3 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.3 | 1.0 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.3 | 1.3 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.3 | 1.6 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.3 | 1.3 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.3 | 1.6 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.3 | 0.6 | GO:0045869 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045869) |
| 0.3 | 0.9 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) |
| 0.3 | 1.5 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.3 | 3.0 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.3 | 2.1 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.3 | 1.8 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.3 | 1.1 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.3 | 1.4 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.3 | 3.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.3 | 1.7 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.3 | 1.3 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.3 | 4.2 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.3 | 1.0 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.3 | 1.0 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.3 | 3.3 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.2 | 0.7 | GO:0046032 | ADP catabolic process(GO:0046032) |
| 0.2 | 1.0 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.2 | 1.0 | GO:0030647 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.2 | 1.9 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.2 | 4.0 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.2 | 1.2 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.2 | 2.6 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.2 | 0.5 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.2 | 1.4 | GO:0098700 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.2 | 4.6 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.2 | 0.9 | GO:0060046 | regulation of acrosome reaction(GO:0060046) |
| 0.2 | 2.0 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.2 | 0.6 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.2 | 3.0 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.2 | 0.6 | GO:1905065 | hematopoietic stem cell migration(GO:0035701) positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.2 | 2.5 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.2 | 0.6 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.2 | 0.8 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.2 | 1.4 | GO:0051461 | protein import into peroxisome matrix, docking(GO:0016560) corticotropin secretion(GO:0051458) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.2 | 1.2 | GO:0009136 | ADP biosynthetic process(GO:0006172) nucleoside diphosphate biosynthetic process(GO:0009133) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) dATP metabolic process(GO:0046060) |
| 0.2 | 1.8 | GO:0046459 | short-chain fatty acid metabolic process(GO:0046459) |
| 0.2 | 1.0 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.2 | 1.2 | GO:1903236 | regulation of leukocyte tethering or rolling(GO:1903236) |
| 0.2 | 0.6 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.2 | 2.6 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.2 | 1.3 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.2 | 1.1 | GO:0001763 | morphogenesis of a branching structure(GO:0001763) |
| 0.2 | 0.8 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.2 | 0.6 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.2 | 0.6 | GO:1903225 | regulation of endodermal cell fate specification(GO:0042663) negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 0.2 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.2 | 1.1 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.2 | 1.3 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.2 | 0.7 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.2 | 0.9 | GO:0061009 | common bile duct development(GO:0061009) |
| 0.2 | 0.9 | GO:0033136 | serine phosphorylation of STAT3 protein(GO:0033136) serine phosphorylation of STAT protein(GO:0042501) |
| 0.2 | 1.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.2 | 8.6 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.2 | 2.0 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.2 | 1.2 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.2 | 1.7 | GO:0031999 | negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.2 | 2.1 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.2 | 1.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.2 | 0.8 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.2 | 0.5 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.2 | 3.5 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 1.9 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) |
| 0.1 | 0.9 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.1 | 1.8 | GO:0002097 | tRNA wobble base modification(GO:0002097) |
| 0.1 | 0.6 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.1 | 0.4 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 1.4 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.1 | 3.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 1.4 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 0.6 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 2.2 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.1 | 0.4 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.1 | 1.3 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 0.9 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.5 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.6 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.4 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.1 | 0.2 | GO:0002922 | positive regulation of humoral immune response(GO:0002922) |
| 0.1 | 1.4 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 0.7 | GO:1903624 | regulation of apoptotic DNA fragmentation(GO:1902510) regulation of DNA catabolic process(GO:1903624) |
| 0.1 | 0.4 | GO:0048352 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) |
| 0.1 | 3.1 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 0.5 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 0.6 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.1 | 2.3 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.1 | 0.2 | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.1 | 2.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.8 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 3.3 | GO:0001947 | heart looping(GO:0001947) |
| 0.1 | 1.0 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 2.4 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 1.0 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 1.1 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 0.4 | GO:0034239 | regulation of macrophage fusion(GO:0034239) |
| 0.1 | 1.0 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.1 | 2.2 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.1 | 1.7 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 3.3 | GO:0000578 | embryonic axis specification(GO:0000578) |
| 0.1 | 2.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 1.2 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 1.7 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 1.0 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.1 | 2.1 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.1 | 1.0 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) |
| 0.1 | 3.2 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 0.3 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 1.6 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.1 | 0.5 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.1 | 2.0 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 0.1 | 2.4 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 0.6 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.6 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 0.2 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.1 | 0.2 | GO:0036509 | trimming of terminal mannose on B branch(GO:0036509) |
| 0.1 | 1.1 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) |
| 0.1 | 1.6 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 | 1.0 | GO:0010388 | cullin deneddylation(GO:0010388) |
| 0.1 | 5.7 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.1 | 0.1 | GO:0060025 | regulation of synaptic activity(GO:0060025) |
| 0.1 | 2.2 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 7.7 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.1 | 2.6 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.1 | 0.6 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 0.6 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.6 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 1.3 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 1.3 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 2.3 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.1 | 1.1 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.1 | 0.9 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 1.1 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 0.2 | GO:2000564 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) CD8-positive, alpha-beta T cell proliferation(GO:0035740) positive regulation of interleukin-12 biosynthetic process(GO:0045084) negative regulation of regulatory T cell differentiation(GO:0045590) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.1 | 0.2 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.1 | 0.2 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.1 | 3.5 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 2.6 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.1 | 0.3 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.1 | 0.3 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.1 | 1.7 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 2.2 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.1 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.4 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 3.3 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.1 | 0.5 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 1.1 | GO:0045909 | positive regulation of vasodilation(GO:0045909) |
| 0.1 | 1.5 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.1 | 0.2 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.3 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) |
| 0.1 | 0.2 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.1 | 0.8 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 0.7 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.1 | 0.4 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 1.3 | GO:0035058 | nonmotile primary cilium assembly(GO:0035058) |
| 0.1 | 0.2 | GO:0032621 | interleukin-18 production(GO:0032621) positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.1 | 0.8 | GO:0007099 | centriole replication(GO:0007099) |
| 0.1 | 5.5 | GO:0000075 | cell cycle checkpoint(GO:0000075) |
| 0.1 | 0.3 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.1 | 0.8 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 | 0.9 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.4 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 1.0 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 1.1 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.6 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0042167 | heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.3 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 1.1 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.3 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.6 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.1 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) |
| 0.0 | 1.0 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.6 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 1.5 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.3 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.6 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 0.4 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.2 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.4 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.0 | 0.2 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 0.2 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.0 | 0.9 | GO:0000060 | protein import into nucleus, translocation(GO:0000060) |
| 0.0 | 2.3 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.4 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.3 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 | 1.0 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.2 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 | 2.0 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 4.1 | GO:0051099 | positive regulation of binding(GO:0051099) |
| 0.0 | 0.8 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.2 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.9 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.0 | 1.2 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 1.0 | GO:0009225 | nucleotide-sugar metabolic process(GO:0009225) |
| 0.0 | 0.5 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 1.1 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 0.6 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:0010560 | positive regulation of glycoprotein biosynthetic process(GO:0010560) |
| 0.0 | 0.5 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.5 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.0 | 0.2 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.4 | GO:0035337 | fatty-acyl-CoA metabolic process(GO:0035337) |
| 0.0 | 1.8 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.7 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.5 | GO:0060416 | response to growth hormone(GO:0060416) |
| 0.0 | 1.1 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.5 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 2.0 | GO:0030518 | intracellular steroid hormone receptor signaling pathway(GO:0030518) |
| 0.0 | 0.2 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.2 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 1.2 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 1.0 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0002834 | immune response to tumor cell(GO:0002418) regulation of response to tumor cell(GO:0002834) regulation of immune response to tumor cell(GO:0002837) |
| 0.0 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.7 | GO:0045580 | regulation of T cell differentiation(GO:0045580) |
| 0.0 | 0.2 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.4 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.3 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 1.8 | GO:0016125 | sterol metabolic process(GO:0016125) |
| 0.0 | 0.1 | GO:0090043 | tubulin deacetylation(GO:0090042) regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.0 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0006582 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.7 | GO:0050868 | negative regulation of T cell activation(GO:0050868) |
| 0.0 | 0.1 | GO:0001711 | endodermal cell fate commitment(GO:0001711) |
| 0.0 | 0.6 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.6 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 0.4 | GO:0070527 | platelet aggregation(GO:0070527) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 23.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 2.0 | 8.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 1.2 | 3.6 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 1.1 | 6.8 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 1.1 | 5.5 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 1.1 | 3.2 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.9 | 2.6 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.8 | 2.5 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.8 | 2.3 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.7 | 7.2 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.7 | 2.1 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.7 | 2.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.7 | 2.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.7 | 6.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.7 | 2.6 | GO:0071942 | XPC complex(GO:0071942) |
| 0.6 | 8.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.6 | 3.0 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.6 | 2.9 | GO:0001652 | granular component(GO:0001652) |
| 0.6 | 2.8 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.5 | 6.0 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.5 | 1.6 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.5 | 7.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.4 | 1.3 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.4 | 2.9 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.4 | 2.3 | GO:0061689 | paranodal junction(GO:0033010) tricellular tight junction(GO:0061689) |
| 0.4 | 1.8 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.4 | 1.4 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.3 | 3.8 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.3 | 1.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.3 | 1.7 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.3 | 2.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.3 | 1.2 | GO:0097413 | Lewy body(GO:0097413) |
| 0.3 | 6.1 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.3 | 0.9 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.3 | 4.6 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.3 | 3.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.3 | 2.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.3 | 2.7 | GO:0000800 | lateral element(GO:0000800) |
| 0.3 | 3.7 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.3 | 1.8 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.3 | 1.0 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.3 | 1.0 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.2 | 3.7 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 1.3 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.2 | 0.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 1.6 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.2 | 2.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.2 | 2.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 1.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 2.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.2 | 2.6 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.2 | 0.7 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.2 | 0.5 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.2 | 1.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 1.0 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 0.9 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.9 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 1.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.8 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 10.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.9 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 1.3 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 0.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 0.7 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.1 | 2.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 0.8 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 1.7 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 1.0 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 2.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 1.0 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 1.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 3.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.7 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 2.0 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 0.8 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.1 | 1.3 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 0.9 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 3.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.7 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 1.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 0.6 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 1.6 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 1.7 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 1.9 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 1.3 | GO:0045120 | pronucleus(GO:0045120) |
| 0.1 | 3.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 1.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 5.6 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 0.2 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 0.7 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 0.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.5 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 2.1 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 2.9 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 1.8 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 0.4 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.5 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 1.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.2 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.1 | 0.2 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.1 | 5.4 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 3.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 5.0 | GO:0005813 | centrosome(GO:0005813) |
| 0.1 | 2.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.2 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 0.9 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 0.3 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 2.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.8 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.0 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.0 | 0.4 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 2.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 4.9 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.8 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.6 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 2.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.4 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 2.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 10.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.2 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 3.8 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 1.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 2.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.5 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 3.2 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 0.6 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.9 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.5 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 1.3 | GO:0022627 | small ribosomal subunit(GO:0015935) cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.5 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 1.0 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 1.0 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.4 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 2.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.2 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 8.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.8 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.5 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.1 | GO:1990462 | omegasome(GO:1990462) |
| 0.0 | 0.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.5 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.6 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.2 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.2 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.8 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 2.0 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.3 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 1.0 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.3 | 21.9 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 5.8 | 23.2 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 2.1 | 6.2 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 1.7 | 8.7 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 1.4 | 8.4 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 1.1 | 4.5 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.9 | 2.8 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.8 | 2.5 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) serine binding(GO:0070905) |
| 0.8 | 6.8 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.7 | 2.2 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.7 | 4.3 | GO:0002135 | CTP binding(GO:0002135) |
| 0.7 | 2.1 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) dinucleotide insertion or deletion binding(GO:0032139) |
| 0.7 | 2.8 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.6 | 2.5 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.6 | 4.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.6 | 1.8 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 0.6 | 3.0 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.6 | 4.8 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.6 | 2.3 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.5 | 1.6 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.5 | 2.0 | GO:0016631 | enoyl-[acyl-carrier-protein] reductase activity(GO:0016631) 2,3-dihydroxy-2,3-dihydro-phenylpropionate dehydrogenase activity(GO:0018498) cis-2,3-dihydrodiol DDT dehydrogenase activity(GO:0018499) trans-9R,10R-dihydrodiolphenanthrene dehydrogenase activity(GO:0018500) cis-chlorobenzene dihydrodiol dehydrogenase activity(GO:0018501) 2,5-dichloro-2,5-cyclohexadiene-1,4-diol dehydrogenase activity(GO:0018502) trans-1,2-dihydrodiolphenanthrene dehydrogenase activity(GO:0018503) 3,4-dihydroxy-3,4-dihydrofluorene dehydrogenase activity(GO:0034790) benzo(a)pyrene-trans-11,12-dihydrodiol dehydrogenase activity(GO:0034805) benzo(a)pyrene-cis-4,5-dihydrodiol dehydrogenase activity(GO:0034809) citronellyl-CoA dehydrogenase activity(GO:0034824) menthone dehydrogenase activity(GO:0034838) phthalate 3,4-cis-dihydrodiol dehydrogenase activity(GO:0034912) cinnamate reductase activity(GO:0043786) NADPH-dependent curcumin reductase activity(GO:0052849) NADPH-dependent dihydrocurcumin reductase activity(GO:0052850) |
| 0.5 | 2.9 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.5 | 2.4 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.5 | 1.8 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.4 | 2.7 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.4 | 7.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.4 | 6.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.4 | 1.6 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.4 | 3.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.4 | 1.2 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.4 | 2.8 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.4 | 4.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.4 | 4.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.4 | 1.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.4 | 1.5 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.4 | 2.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.3 | 1.4 | GO:0036468 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 0.3 | 1.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.3 | 1.5 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.3 | 2.4 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.3 | 0.6 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.3 | 2.1 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.3 | 2.9 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.3 | 2.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.3 | 1.4 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.3 | 4.0 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.3 | 0.8 | GO:0018738 | S-formylglutathione hydrolase activity(GO:0018738) |
| 0.3 | 2.5 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.3 | 2.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.3 | 1.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.3 | 1.6 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.3 | 1.6 | GO:0019841 | retinol binding(GO:0019841) |
| 0.3 | 1.0 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.3 | 1.3 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.2 | 2.0 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.2 | 1.2 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.2 | 5.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 1.0 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.2 | 3.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 3.4 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.2 | 1.6 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 1.7 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.2 | 1.9 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 2.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 1.7 | GO:0008251 | tRNA-specific adenosine deaminase activity(GO:0008251) |
| 0.2 | 0.8 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.2 | 0.6 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.2 | 0.6 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.2 | 0.6 | GO:0019863 | IgE binding(GO:0019863) |
| 0.2 | 4.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.2 | 0.8 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.2 | 6.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.2 | 1.3 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.2 | 1.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 2.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 0.6 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.2 | 3.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.2 | 0.7 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.2 | 3.8 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.2 | 1.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.2 | 1.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.2 | 0.7 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.2 | 0.5 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.2 | 0.9 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 0.9 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.2 | 0.7 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 1.0 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.2 | 1.3 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.2 | 1.3 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 3.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 0.5 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.2 | 1.6 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.2 | 2.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.2 | 0.6 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.2 | 4.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.2 | 1.4 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.2 | 1.2 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.2 | 1.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 0.5 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.1 | 4.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 1.4 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 2.4 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 1.6 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 2.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 2.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.4 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.1 | 1.6 | GO:0048038 | quinone binding(GO:0048038) |
| 0.1 | 1.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.4 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 0.7 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 3.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 1.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.5 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.5 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 1.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 7.1 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 0.8 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 2.2 | GO:0052890 | oxidoreductase activity, acting on the CH-CH group of donors, with a flavin as acceptor(GO:0052890) |
| 0.1 | 0.8 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 0.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 1.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 1.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 0.7 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 0.5 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 2.2 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 5.9 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.1 | 2.3 | GO:0034061 | DNA-directed DNA polymerase activity(GO:0003887) DNA polymerase activity(GO:0034061) |
| 0.1 | 5.2 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 1.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.7 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 2.1 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 1.6 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.1 | 0.8 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 4.1 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.1 | 0.2 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 0.4 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.1 | 0.8 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 1.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 1.0 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 1.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.2 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 2.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 1.9 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 0.3 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.1 | 0.6 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 1.3 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.3 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.1 | 1.3 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.1 | 0.3 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.4 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.1 | 1.0 | GO:0042556 | cobinamide kinase activity(GO:0008819) phytol kinase activity(GO:0010276) phenol kinase activity(GO:0018720) cyclin-dependent protein kinase activating kinase regulator activity(GO:0019914) inositol tetrakisphosphate 2-kinase activity(GO:0032942) heptose 7-phosphate kinase activity(GO:0033785) aminoglycoside phosphotransferase activity(GO:0034071) eukaryotic elongation factor-2 kinase regulator activity(GO:0042556) eukaryotic elongation factor-2 kinase activator activity(GO:0042557) LPPG:FO 2-phospho-L-lactate transferase activity(GO:0043743) cytidine kinase activity(GO:0043771) glycerate 2-kinase activity(GO:0043798) (S)-lactate 2-kinase activity(GO:0043841) phosphoserine:homoserine phosphotransferase activity(GO:0043899) L-seryl-tRNA(Sec) kinase activity(GO:0043915) phosphocholine transferase activity(GO:0044605) GTP-dependent polynucleotide kinase activity(GO:0051735) farnesol kinase activity(GO:0052668) CTP:2-trans,-6-trans-farnesol kinase activity(GO:0052669) geraniol kinase activity(GO:0052670) geranylgeraniol kinase activity(GO:0052671) CTP:geranylgeraniol kinase activity(GO:0052672) prenol kinase activity(GO:0052673) 1-phosphatidylinositol-5-kinase activity(GO:0052810) 1-phosphatidylinositol-3-phosphate 4-kinase activity(GO:0052811) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.1 | 2.2 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 0.1 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) |
| 0.1 | 1.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 3.6 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.1 | 0.5 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 0.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 4.1 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.1 | 0.8 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 0.7 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 0.2 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 4.9 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 1.0 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 2.4 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 0.3 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.8 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.4 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 1.7 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.2 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.2 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.0 | 0.3 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.3 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 1.3 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 1.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.9 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.0 | GO:0034930 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) heparan sulfate 2-O-sulfotransferase activity(GO:0004394) HNK-1 sulfotransferase activity(GO:0016232) heparan sulfate 6-O-sulfotransferase activity(GO:0017095) trans-9R,10R-dihydrodiolphenanthrene sulfotransferase activity(GO:0018721) 1-phenanthrol sulfotransferase activity(GO:0018722) 3-phenanthrol sulfotransferase activity(GO:0018723) 4-phenanthrol sulfotransferase activity(GO:0018724) trans-3,4-dihydrodiolphenanthrene sulfotransferase activity(GO:0018725) 9-phenanthrol sulfotransferase activity(GO:0018726) 2-phenanthrol sulfotransferase activity(GO:0018727) phenanthrol sulfotransferase activity(GO:0019111) 1-hydroxypyrene sulfotransferase activity(GO:0034930) proteoglycan sulfotransferase activity(GO:0050698) cholesterol sulfotransferase activity(GO:0051922) hydroxyjasmonate sulfotransferase activity(GO:0080131) |
| 0.0 | 0.3 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 1.5 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.0 | 0.2 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 1.1 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 1.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.4 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) |
| 0.0 | 0.9 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.6 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.7 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.2 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.0 | 1.9 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 2.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.6 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 3.4 | GO:0043774 | UDP-N-acetylmuramoylalanyl-D-glutamyl-2,6-diaminopimelate-D-alanyl-D-alanine ligase activity(GO:0008766) ribosomal S6-glutamic acid ligase activity(GO:0018169) coenzyme F420-0 gamma-glutamyl ligase activity(GO:0043773) coenzyme F420-2 alpha-glutamyl ligase activity(GO:0043774) protein-glycine ligase activity(GO:0070735) protein-glycine ligase activity, initiating(GO:0070736) protein-glycine ligase activity, elongating(GO:0070737) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 0.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.2 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.8 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.2 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.0 | 0.7 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 3.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.2 | GO:0016835 | carbon-oxygen lyase activity(GO:0016835) |
| 0.0 | 1.0 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 1.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.0 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 1.0 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.6 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.5 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 1.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.2 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 4.3 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 14.4 | GO:0044822 | poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.2 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.2 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.1 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |