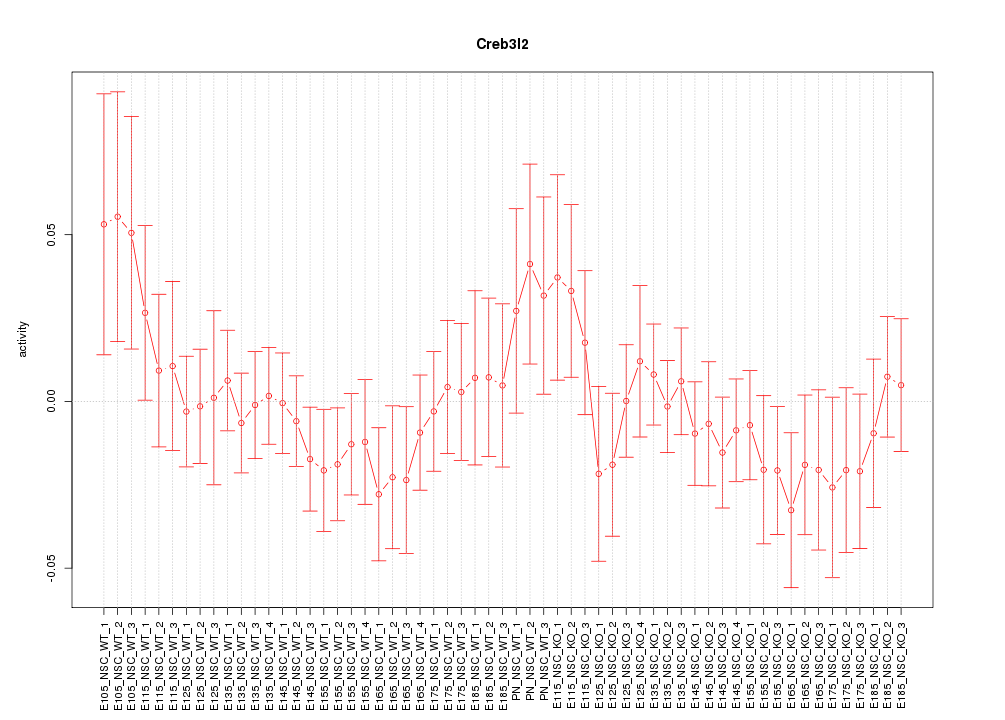
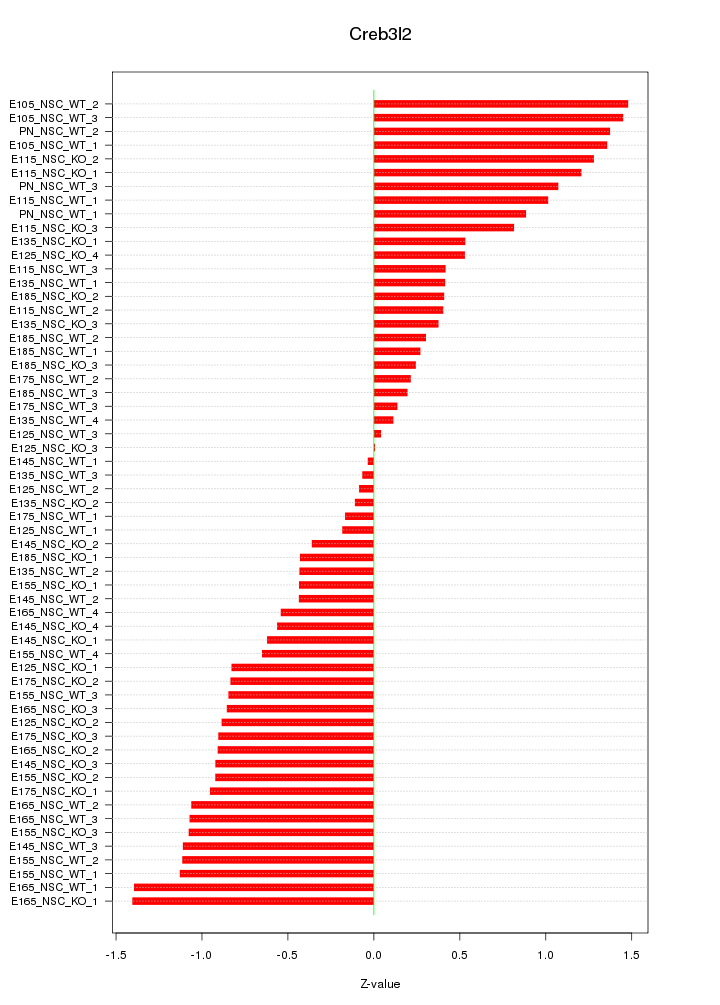
Motif ID: Creb3l2
Z-value: 0.801

Transcription factors associated with Creb3l2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Creb3l2 | ENSMUSG00000038648.5 | Creb3l2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Creb3l2 | mm10_v2_chr6_-_37442095_37442154 | 0.31 | 1.9e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.0 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 1.4 | 20.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 1.3 | 5.2 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 1.1 | 5.7 | GO:0072429 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 1.0 | 4.0 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.8 | 2.4 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.8 | 12.6 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.8 | 2.3 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.8 | 3.8 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.7 | 3.6 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.7 | 2.8 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.6 | 3.9 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.4 | 4.4 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.4 | 5.2 | GO:0014842 | regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.4 | 2.9 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.4 | 2.1 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.3 | 1.0 | GO:0045359 | positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.3 | 1.9 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.3 | 0.9 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.3 | 0.9 | GO:2000813 | actin filament uncapping(GO:0051695) negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.3 | 2.2 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.3 | 0.8 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 0.3 | 4.4 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.2 | 4.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.2 | 0.9 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.2 | 1.2 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.2 | 1.8 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.2 | 0.6 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.2 | 7.8 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.2 | 13.5 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.2 | 1.8 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.2 | 1.3 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.2 | 3.1 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.2 | 1.7 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.2 | 0.9 | GO:1903887 | motile primary cilium assembly(GO:1903887) |
| 0.2 | 0.8 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.2 | 0.8 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 | 0.8 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 3.9 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.1 | 1.8 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.1 | 1.2 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 | 1.1 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 1.2 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 0.8 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.1 | 3.9 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.1 | 0.3 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 3.4 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 | 0.4 | GO:1904451 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.1 | 0.8 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 3.1 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 0.3 | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.1 | 0.6 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 2.1 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.1 | 2.6 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.1 | 0.9 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 1.0 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 2.5 | GO:0006739 | NADP metabolic process(GO:0006739) |
| 0.1 | 0.2 | GO:0070625 | zymogen granule exocytosis(GO:0070625) |
| 0.1 | 1.5 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.7 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 3.0 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.3 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.2 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.4 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 | 2.5 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 4.7 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 3.6 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 1.2 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 1.6 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.4 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 5.3 | GO:0043010 | camera-type eye development(GO:0043010) |
| 0.0 | 1.5 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 0.3 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.0 | 1.2 | GO:0048041 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) |
| 0.0 | 3.0 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 0.9 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.0 | 0.3 | GO:0048255 | mRNA stabilization(GO:0048255) |
| 0.0 | 0.5 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.2 | GO:0005980 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.5 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.7 | GO:0055072 | iron ion homeostasis(GO:0055072) |
| 0.0 | 0.5 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.2 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.7 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.9 | 3.6 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.8 | 3.9 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.6 | 1.8 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.6 | 4.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.5 | 2.9 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.4 | 1.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.3 | 4.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.3 | 2.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.2 | 4.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.2 | 2.1 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.2 | 0.8 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.2 | 3.0 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 1.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.9 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.1 | 1.0 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.1 | 1.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.9 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 0.4 | GO:1990795 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) |
| 0.1 | 0.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.4 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.1 | 0.7 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 5.2 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.9 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 19.1 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 2.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 1.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 3.5 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 3.5 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.0 | 10.3 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.6 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.9 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 5.7 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 17.1 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 1.5 | GO:0016607 | nuclear speck(GO:0016607) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 20.3 | GO:0018121 | imidazoleglycerol-phosphate synthase activity(GO:0000107) NAD(P)-cysteine ADP-ribosyltransferase activity(GO:0018071) NAD(P)-asparagine ADP-ribosyltransferase activity(GO:0018121) NAD(P)-serine ADP-ribosyltransferase activity(GO:0018127) 7-cyano-7-deazaguanine tRNA-ribosyltransferase activity(GO:0043867) purine deoxyribosyltransferase activity(GO:0044102) |
| 1.3 | 5.2 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 1.2 | 3.6 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 1.0 | 3.9 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.9 | 2.8 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.9 | 2.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.8 | 2.4 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.7 | 2.9 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.5 | 4.0 | GO:0103116 | alpha-D-galactofuranose transporter activity(GO:0103116) steroid hormone binding(GO:1990239) |
| 0.5 | 13.5 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.5 | 1.8 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.4 | 1.2 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.3 | 4.4 | GO:0005536 | glucose binding(GO:0005536) |
| 0.3 | 1.5 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.3 | 0.8 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.3 | 4.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.3 | 0.8 | GO:0017084 | glutamate 5-kinase activity(GO:0004349) glutamate-5-semialdehyde dehydrogenase activity(GO:0004350) delta1-pyrroline-5-carboxylate synthetase activity(GO:0017084) amino acid kinase activity(GO:0019202) |
| 0.2 | 3.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.2 | 1.6 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 0.6 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.2 | 3.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.2 | 7.8 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.2 | 2.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 2.3 | GO:0048038 | quinone binding(GO:0048038) |
| 0.2 | 2.8 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 2.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 1.7 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.2 | 1.8 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.2 | 1.9 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 4.4 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.2 | 0.8 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.2 | 0.8 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.2 | 2.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 1.5 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 1.3 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) activin binding(GO:0048185) |
| 0.1 | 2.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.6 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 0.6 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 0.3 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 5.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 2.5 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 1.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.7 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 3.0 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 2.5 | GO:0050661 | NADP binding(GO:0050661) |
| 0.1 | 0.8 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 2.8 | GO:0004120 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.1 | 0.9 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.1 | 3.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 3.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.2 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) |
| 0.0 | 0.2 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 5.7 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 3.9 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 10.8 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 3.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 3.9 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 2.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 5.2 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.3 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 5.1 | GO:0005198 | structural molecule activity(GO:0005198) |