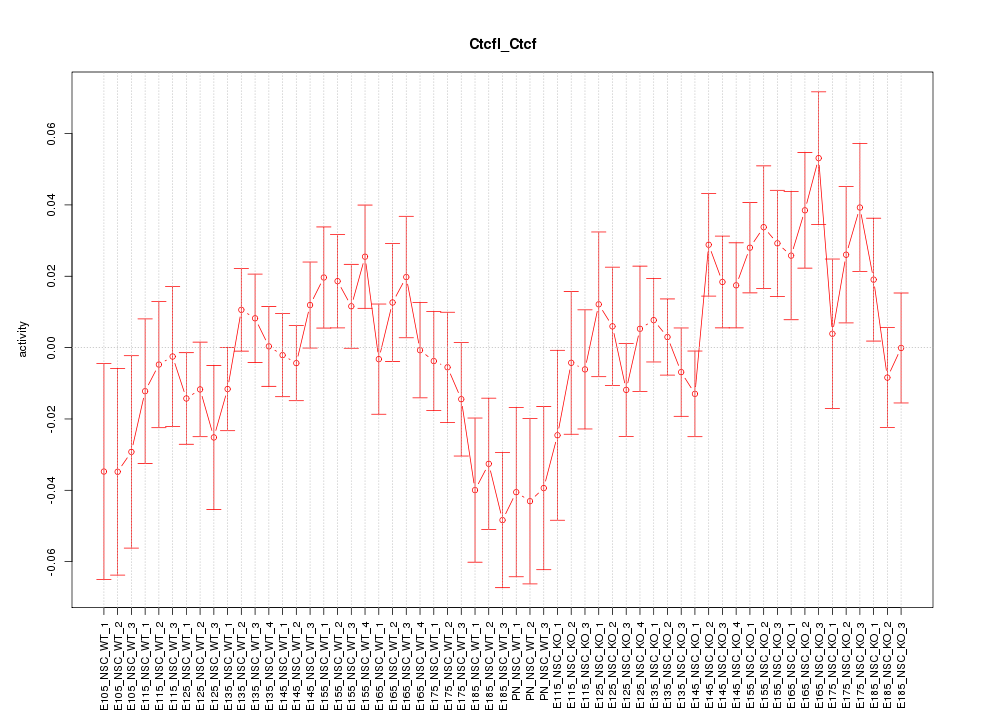
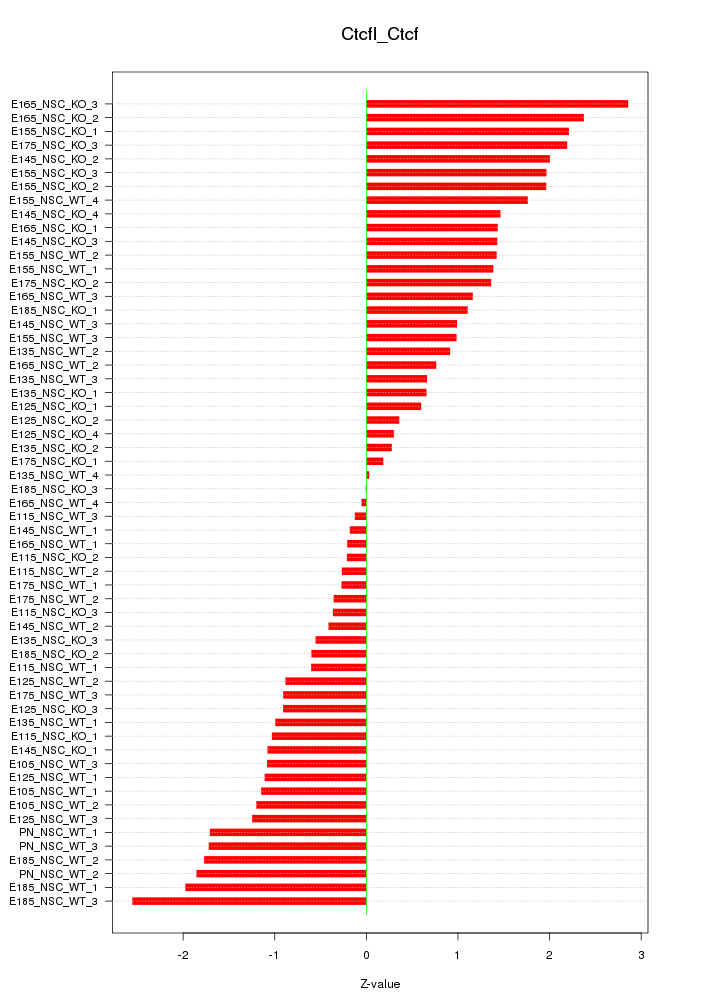
Motif ID: Ctcfl_Ctcf
Z-value: 1.268
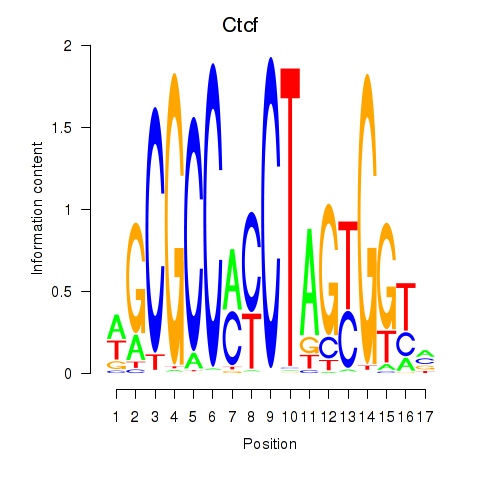
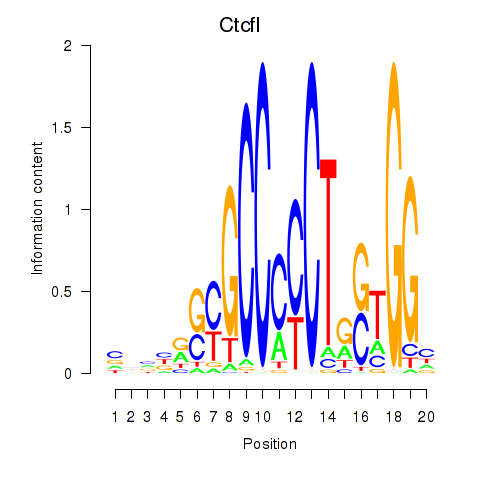
Transcription factors associated with Ctcfl_Ctcf:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Ctcf | ENSMUSG00000005698.9 | Ctcf |
| Ctcfl | ENSMUSG00000070495.5 | Ctcfl |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ctcfl | mm10_v2_chr2_-_173119402_173119525 | -0.26 | 4.5e-02 | Click! |
| Ctcf | mm10_v2_chr8_+_105636509_105636589 | -0.20 | 1.3e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 14.6 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 3.0 | 9.1 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 2.7 | 10.7 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 2.5 | 7.5 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 2.4 | 11.9 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 2.4 | 7.1 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 2.3 | 18.1 | GO:1903423 | positive regulation of synaptic vesicle recycling(GO:1903423) |
| 2.2 | 34.6 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 2.1 | 10.4 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 2.1 | 6.2 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 1.9 | 7.6 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 1.9 | 5.6 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 1.8 | 7.2 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 1.3 | 6.6 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 1.3 | 5.2 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) |
| 1.3 | 9.0 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 1.2 | 3.6 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 1.2 | 3.5 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 1.1 | 3.4 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 1.0 | 3.0 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 1.0 | 6.0 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 1.0 | 4.0 | GO:0035624 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 1.0 | 3.9 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.9 | 2.8 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.9 | 3.7 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.8 | 2.5 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.8 | 2.5 | GO:0060023 | soft palate development(GO:0060023) |
| 0.8 | 1.5 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.7 | 10.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.7 | 5.7 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.7 | 2.1 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.7 | 2.1 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.7 | 2.0 | GO:0009838 | abscission(GO:0009838) positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.6 | 1.9 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.6 | 1.9 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.6 | 1.8 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.6 | 1.8 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.6 | 3.0 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.6 | 3.9 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.6 | 1.7 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.6 | 5.0 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.5 | 5.5 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.5 | 1.6 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.5 | 8.1 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.5 | 3.0 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.5 | 0.5 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.5 | 0.9 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.5 | 1.4 | GO:0072425 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.4 | 0.9 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.4 | 1.7 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.4 | 9.0 | GO:0042297 | vocal learning(GO:0042297) imitative learning(GO:0098596) learned vocalization behavior or vocal learning(GO:0098598) |
| 0.4 | 2.9 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.4 | 2.9 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.4 | 2.0 | GO:0072429 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.4 | 3.5 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.4 | 3.1 | GO:0030432 | peristalsis(GO:0030432) |
| 0.4 | 7.4 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.4 | 2.7 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.4 | 3.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.4 | 2.6 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.4 | 2.6 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.4 | 3.7 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.4 | 3.3 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.4 | 1.1 | GO:0070428 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.4 | 1.8 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.4 | 2.8 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.4 | 0.7 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.4 | 4.2 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.3 | 10.8 | GO:0061098 | positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.3 | 4.1 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.3 | 3.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.3 | 2.3 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.3 | 1.6 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.3 | 6.8 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.3 | 1.8 | GO:0045988 | negative regulation of striated muscle contraction(GO:0045988) |
| 0.3 | 2.3 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.3 | 1.4 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.3 | 3.4 | GO:0035898 | parathyroid hormone secretion(GO:0035898) |
| 0.3 | 1.7 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.3 | 1.3 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.3 | 1.3 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.3 | 4.4 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.2 | 1.1 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.2 | 2.5 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.2 | 1.6 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.2 | 5.9 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.2 | 3.2 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.2 | 1.7 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.2 | 0.6 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.2 | 0.6 | GO:1904017 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.2 | 7.2 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.2 | 0.4 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.2 | 0.6 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) peptidyl-glycine modification(GO:0018201) |
| 0.2 | 14.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.2 | 2.1 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.2 | 0.8 | GO:0060266 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) negative regulation of respiratory burst(GO:0060268) |
| 0.2 | 6.3 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.2 | 1.0 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.2 | 5.2 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.2 | 14.3 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.2 | 1.0 | GO:0003383 | apical constriction(GO:0003383) |
| 0.2 | 0.7 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.2 | 2.0 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 3.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 0.2 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.2 | 15.4 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.2 | 1.2 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.2 | 1.7 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.2 | 3.2 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.2 | 0.8 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.1 | 0.7 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 12.3 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.1 | 1.3 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.1 | 0.7 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 0.4 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 0.8 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 0.9 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 2.1 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.1 | 2.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 2.0 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 5.3 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 0.5 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.9 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 | 2.2 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 1.9 | GO:1903861 | regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 0.9 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 1.3 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.1 | 1.0 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.1 | 2.4 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 0.8 | GO:0071816 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 0.4 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 2.1 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 0.9 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 2.5 | GO:0015991 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.1 | 2.1 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 1.3 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.1 | 1.5 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 7.6 | GO:1903749 | positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.1 | 1.5 | GO:0014898 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.1 | 1.9 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.1 | 0.7 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 | 3.3 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 0.4 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.1 | 1.1 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 1.4 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.6 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 0.7 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 0.4 | GO:0032026 | response to magnesium ion(GO:0032026) |
| 0.1 | 1.6 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.1 | 1.7 | GO:0045620 | negative regulation of lymphocyte differentiation(GO:0045620) |
| 0.1 | 0.7 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.1 | 2.0 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.1 | 0.9 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.1 | 4.1 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.1 | 0.3 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.1 | 1.1 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.1 | 0.5 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.1 | 0.9 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.1 | 0.2 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) peptidyl-arginine omega-N-methylation(GO:0035247) |
| 0.1 | 1.3 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.4 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 1.2 | GO:0042755 | eating behavior(GO:0042755) |
| 0.0 | 7.1 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.8 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.6 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.8 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 0.0 | 0.1 | GO:0001705 | ectoderm formation(GO:0001705) |
| 0.0 | 0.6 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 1.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.7 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 3.2 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 2.4 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 1.4 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.0 | 1.2 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 2.4 | GO:0000725 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 | 2.1 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.0 | 1.6 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.1 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.9 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.6 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.1 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.0 | 2.4 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.4 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.8 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 1.1 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.1 | GO:0019478 | D-amino acid catabolic process(GO:0019478) |
| 0.0 | 1.1 | GO:0009612 | response to mechanical stimulus(GO:0009612) |
| 0.0 | 0.5 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.5 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 0.4 | GO:0070266 | necroptotic process(GO:0070266) |
| 0.0 | 0.5 | GO:0061036 | positive regulation of cartilage development(GO:0061036) |
| 0.0 | 0.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 1.1 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.2 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.1 | GO:0015675 | nickel cation transport(GO:0015675) |
| 0.0 | 0.3 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.0 | 0.3 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 30.7 | GO:0045298 | tubulin complex(GO:0045298) |
| 2.2 | 11.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 1.8 | 29.0 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 1.4 | 20.4 | GO:0043196 | varicosity(GO:0043196) |
| 1.1 | 5.7 | GO:0005683 | U7 snRNP(GO:0005683) |
| 1.1 | 5.6 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 1.1 | 3.2 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 1.0 | 3.0 | GO:1990075 | kinesin II complex(GO:0016939) periciliary membrane compartment(GO:1990075) |
| 0.7 | 3.7 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.7 | 8.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.7 | 6.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.6 | 1.8 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.6 | 5.5 | GO:0005883 | neurofilament(GO:0005883) |
| 0.6 | 4.7 | GO:0097433 | dense body(GO:0097433) |
| 0.5 | 7.4 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.5 | 5.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.5 | 1.9 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.5 | 3.7 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.5 | 1.4 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.5 | 4.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.5 | 2.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.4 | 4.0 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.4 | 2.2 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.4 | 6.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.4 | 2.6 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.4 | 2.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.4 | 0.8 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.4 | 2.0 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.4 | 3.3 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.3 | 3.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.3 | 0.3 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.3 | 3.0 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.3 | 2.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.3 | 0.9 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.3 | 2.0 | GO:0090543 | Flemming body(GO:0090543) |
| 0.3 | 5.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.3 | 25.9 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.2 | 6.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 9.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 1.1 | GO:0097361 | CIA complex(GO:0097361) |
| 0.2 | 2.5 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.2 | 1.8 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.2 | 2.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.2 | 2.6 | GO:0001527 | microfibril(GO:0001527) |
| 0.2 | 2.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.2 | 0.8 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.2 | 0.4 | GO:0031523 | Myb complex(GO:0031523) |
| 0.2 | 5.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 2.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.2 | 3.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 9.6 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.2 | 7.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.2 | 1.6 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.2 | 26.9 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.2 | 3.8 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 1.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 2.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 1.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.7 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.9 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 3.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 1.6 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.6 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 2.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 4.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 0.7 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 1.1 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.1 | 4.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 2.1 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 1.8 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 14.7 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 0.2 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 2.8 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 0.1 | 1.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 1.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 3.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 6.4 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 16.5 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.1 | 11.2 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 5.2 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 2.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 1.5 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 1.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.6 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 2.7 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 1.7 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 4.2 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 1.2 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.0 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.2 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 0.4 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.0 | 6.1 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.1 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 1.5 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.6 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 18.5 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 2.6 | 25.9 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 2.6 | 10.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 2.5 | 7.6 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 2.3 | 9.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 2.1 | 10.4 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 2.1 | 2.1 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 2.1 | 6.2 | GO:0032450 | oligo-1,6-glucosidase activity(GO:0004574) maltose alpha-glucosidase activity(GO:0032450) |
| 1.9 | 5.7 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 1.5 | 29.0 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 1.3 | 30.7 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 1.3 | 4.0 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) epinephrine binding(GO:0051379) |
| 1.3 | 3.8 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 1.0 | 6.0 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 1.0 | 3.9 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.9 | 2.8 | GO:0035175 | histone kinase activity (H3-S10 specific)(GO:0035175) |
| 0.9 | 6.8 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.7 | 2.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.6 | 2.5 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.6 | 1.8 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.6 | 1.8 | GO:0031755 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.6 | 2.9 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.6 | 2.3 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.6 | 5.6 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.5 | 2.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.5 | 4.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.5 | 1.6 | GO:0015173 | hydrogen:amino acid symporter activity(GO:0005280) aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.5 | 3.1 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.5 | 1.9 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.5 | 3.7 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.5 | 4.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.5 | 1.8 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.5 | 18.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.5 | 1.8 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.5 | 2.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.4 | 2.7 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.4 | 1.9 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.4 | 5.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.4 | 3.0 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.4 | 4.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.4 | 2.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.4 | 1.1 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.3 | 3.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.3 | 2.0 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.3 | 3.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.3 | 7.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.3 | 2.8 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.3 | 5.9 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.3 | 1.8 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.3 | 0.9 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.3 | 2.0 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.3 | 3.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.3 | 1.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.3 | 1.6 | GO:0043426 | MRF binding(GO:0043426) |
| 0.2 | 4.7 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.2 | 1.7 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.2 | 0.7 | GO:0004980 | melanocortin receptor activity(GO:0004977) melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.2 | 1.7 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.2 | 1.7 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.2 | 1.9 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.2 | 11.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.2 | 2.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 0.6 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.2 | 3.4 | GO:0050253 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) retinyl-palmitate esterase activity(GO:0050253) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.2 | 3.5 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.2 | 0.7 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.2 | 2.1 | GO:0016443 | bidentate ribonuclease III activity(GO:0016443) |
| 0.2 | 0.5 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.2 | 0.6 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.1 | 7.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.9 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.1 | 5.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 1.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.7 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.1 | 3.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 4.1 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.1 | 1.0 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 2.4 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 1.9 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 4.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 13.6 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.2 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 3.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.8 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 1.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 2.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 3.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 2.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 2.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 2.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 3.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 7.4 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.1 | 0.5 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 6.5 | GO:0005496 | steroid binding(GO:0005496) |
| 0.1 | 0.5 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 3.3 | GO:0045502 | dynein binding(GO:0045502) |
| 0.1 | 0.3 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 0.4 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.2 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 0.3 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.1 | 1.6 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 4.2 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 0.3 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.1 | 8.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 0.3 | GO:0004954 | icosanoid receptor activity(GO:0004953) prostanoid receptor activity(GO:0004954) prostaglandin receptor activity(GO:0004955) |
| 0.1 | 0.9 | GO:0023026 | MHC protein complex binding(GO:0023023) MHC class II protein complex binding(GO:0023026) |
| 0.1 | 0.4 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.8 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 4.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 4.4 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 4.1 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.1 | 1.9 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 0.4 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 1.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 2.7 | GO:0044824 | integrase activity(GO:0008907) T/G mismatch-specific endonuclease activity(GO:0043765) retroviral integrase activity(GO:0044823) retroviral 3' processing activity(GO:0044824) |
| 0.0 | 2.6 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 3.4 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 3.2 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.0 | 2.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 1.9 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 6.8 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 1.0 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.8 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.6 | GO:0004532 | 3'-5'-exoribonuclease activity(GO:0000175) exoribonuclease activity(GO:0004532) exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.0 | 0.4 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 2.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0015099 | nickel cation transmembrane transporter activity(GO:0015099) |
| 0.0 | 0.4 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.1 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.7 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.4 | GO:0008748 | N-ethylmaleimide reductase activity(GO:0008748) reduced coenzyme F420 dehydrogenase activity(GO:0043738) sulfur oxygenase reductase activity(GO:0043826) malolactic enzyme activity(GO:0043883) epoxyqueuosine reductase activity(GO:0052693) |
| 0.0 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.0 | GO:0016778 | diphosphotransferase activity(GO:0016778) |