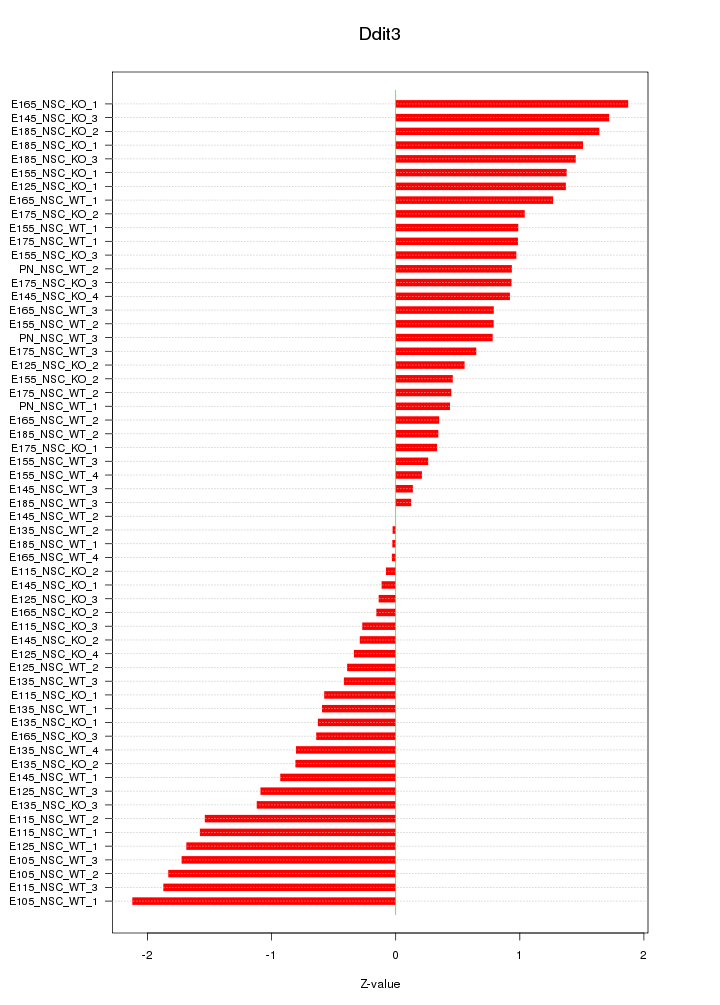
Motif ID: Ddit3
Z-value: 0.991

Transcription factors associated with Ddit3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Ddit3 | ENSMUSG00000025408.9 | Ddit3 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ddit3 | mm10_v2_chr10_+_127290774_127290803 | 0.41 | 1.5e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 11.3 | GO:1902847 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) |
| 1.7 | 8.3 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 1.6 | 4.8 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 1.6 | 14.0 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 1.4 | 4.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 1.1 | 3.2 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 1.1 | 3.2 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.9 | 6.4 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.8 | 4.2 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.8 | 6.5 | GO:0071732 | cellular response to nitric oxide(GO:0071732) |
| 0.7 | 7.4 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.7 | 7.3 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.7 | 2.1 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.6 | 2.4 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.6 | 1.8 | GO:0070649 | meiotic chromosome movement towards spindle pole(GO:0016344) formin-nucleated actin cable assembly(GO:0070649) |
| 0.6 | 3.5 | GO:1903056 | regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.6 | 1.7 | GO:0086042 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) |
| 0.5 | 2.2 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.5 | 3.8 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.5 | 2.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.5 | 2.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.4 | 1.8 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.4 | 1.3 | GO:0030070 | insulin processing(GO:0030070) |
| 0.4 | 2.6 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.4 | 1.3 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.4 | 2.5 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.4 | 1.2 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 0.4 | 1.2 | GO:0042275 | error-free postreplication DNA repair(GO:0042275) |
| 0.4 | 4.8 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.4 | 1.4 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.3 | 6.7 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.3 | 2.4 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.3 | 3.6 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.2 | 3.4 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.2 | 1.2 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.2 | 1.8 | GO:0031424 | keratinization(GO:0031424) |
| 0.2 | 1.8 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.2 | 2.0 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.2 | 1.1 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.2 | 4.4 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.2 | 4.9 | GO:0009409 | response to cold(GO:0009409) |
| 0.2 | 3.4 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.2 | 1.5 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.2 | 1.2 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.1 | 3.6 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 0.9 | GO:0098543 | detection of bacterium(GO:0016045) negative regulation of natural killer cell activation(GO:0032815) detection of other organism(GO:0098543) |
| 0.1 | 0.8 | GO:0071816 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 1.9 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 1.6 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.1 | 2.1 | GO:0048741 | skeletal muscle fiber development(GO:0048741) |
| 0.1 | 0.6 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.1 | 1.0 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 7.0 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.1 | 2.4 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 0.8 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 9.8 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.1 | 2.3 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 1.7 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 | 0.4 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 2.0 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 1.7 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.7 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 0.5 | GO:0051974 | negative regulation of telomerase activity(GO:0051974) |
| 0.0 | 0.7 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 3.3 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.0 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.0 | 0.1 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.4 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.1 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 | 0.1 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 11.3 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 1.0 | 8.3 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.6 | 14.0 | GO:0031430 | M band(GO:0031430) |
| 0.5 | 7.4 | GO:0043203 | axon hillock(GO:0043203) |
| 0.5 | 10.5 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.5 | 1.9 | GO:0031673 | H zone(GO:0031673) |
| 0.5 | 1.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.4 | 4.9 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.4 | 1.2 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.4 | 1.2 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.4 | 14.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.3 | 3.6 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.3 | 4.4 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.2 | 1.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.2 | 7.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 4.8 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.2 | 0.5 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 2.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.4 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 2.5 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 3.6 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 1.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.6 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 2.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 1.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.4 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 1.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 2.1 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 2.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.7 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 5.5 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 7.6 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 3.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 3.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 5.8 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 2.7 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 1.8 | GO:0005938 | cell cortex(GO:0005938) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 1.3 | 6.7 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 1.3 | 3.8 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.8 | 3.9 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.6 | 2.5 | GO:0004104 | cholinesterase activity(GO:0004104) choline binding(GO:0033265) |
| 0.6 | 2.4 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.5 | 11.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.5 | 1.4 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) |
| 0.5 | 14.0 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.4 | 7.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.3 | 2.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.3 | 1.7 | GO:0086080 | connexin binding(GO:0071253) protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.2 | 6.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 0.9 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.2 | 7.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.2 | 3.3 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.2 | 0.6 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.2 | 3.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 4.8 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 4.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) cobalt ion binding(GO:0050897) |
| 0.1 | 3.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 2.0 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 3.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 1.5 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 2.0 | GO:0035380 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) mevaldate reductase activity(GO:0004495) gluconate dehydrogenase activity(GO:0008875) epoxide dehydrogenase activity(GO:0018451) 5-exo-hydroxycamphor dehydrogenase activity(GO:0018452) 2-hydroxytetrahydrofuran dehydrogenase activity(GO:0018453) acetoin dehydrogenase activity(GO:0019152) phenylcoumaran benzylic ether reductase activity(GO:0032442) D-xylose:NADP reductase activity(GO:0032866) L-arabinose:NADP reductase activity(GO:0032867) D-arabinitol dehydrogenase, D-ribulose forming (NADP+) activity(GO:0033709) (R)-(-)-1,2,3,4-tetrahydronaphthol dehydrogenase activity(GO:0034831) 3-hydroxymenthone dehydrogenase activity(GO:0034840) very long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0035380) dihydrotestosterone 17-beta-dehydrogenase activity(GO:0035410) (R)-2-hydroxyisocaproate dehydrogenase activity(GO:0043713) L-arabinose 1-dehydrogenase (NADP+) activity(GO:0044103) L-xylulose reductase (NAD+) activity(GO:0044105) 3-ketoglucose-reductase activity(GO:0048258) D-arabinitol dehydrogenase, D-xylulose forming (NADP+) activity(GO:0052677) |
| 0.1 | 5.5 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.1 | 3.6 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.1 | 2.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 3.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 3.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 1.0 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.1 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 2.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.5 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 2.2 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 1.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 1.6 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 1.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 3.4 | GO:0016810 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds(GO:0016810) |
| 0.0 | 1.2 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 2.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 1.2 | GO:0044325 | ion channel binding(GO:0044325) |