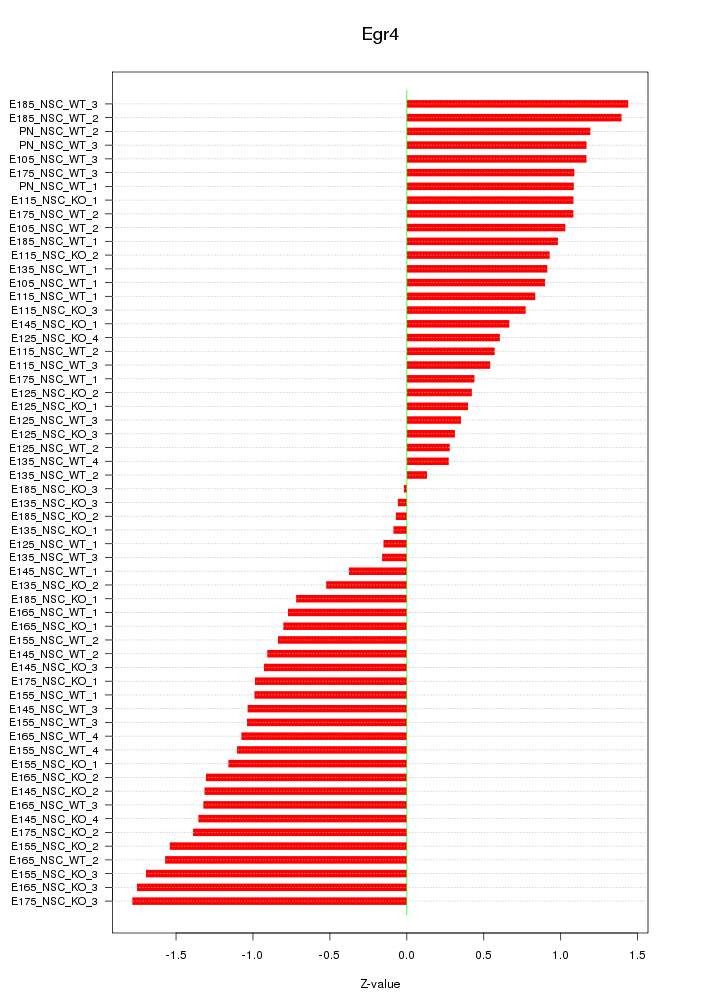
Motif ID: Egr4
Z-value: 0.978

Transcription factors associated with Egr4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Egr4 | ENSMUSG00000071341.3 | Egr4 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 1.1 | 4.4 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 1.1 | 3.2 | GO:0003100 | angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) regulation of systemic arterial blood pressure by endothelin(GO:0003100) beta selection(GO:0043366) |
| 0.9 | 6.4 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.8 | 12.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.7 | 5.0 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.7 | 3.5 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.7 | 2.1 | GO:0045014 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.6 | 2.6 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.6 | 2.3 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.6 | 1.7 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.5 | 2.7 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.5 | 7.5 | GO:0030238 | male sex determination(GO:0030238) |
| 0.5 | 3.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.4 | 10.1 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.4 | 1.2 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.4 | 2.9 | GO:0000050 | urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.3 | 4.2 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.3 | 1.2 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.3 | 1.5 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.3 | 1.2 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.3 | 2.4 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.3 | 5.0 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.3 | 2.9 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.2 | 2.2 | GO:1903624 | regulation of apoptotic DNA fragmentation(GO:1902510) regulation of DNA catabolic process(GO:1903624) |
| 0.2 | 1.1 | GO:0010359 | regulation of anion channel activity(GO:0010359) |
| 0.2 | 1.2 | GO:1900016 | negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.2 | 0.7 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.2 | 2.1 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.2 | 3.7 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.2 | 0.9 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 4.3 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 3.2 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.1 | 4.4 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 1.8 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.1 | 5.9 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 6.1 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.1 | 0.3 | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.1 | 3.0 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 6.2 | GO:0070613 | regulation of protein processing(GO:0070613) |
| 0.1 | 0.9 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 1.7 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.1 | 5.8 | GO:0007601 | visual perception(GO:0007601) |
| 0.1 | 2.9 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 1.5 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.3 | GO:1904754 | positive regulation of vascular associated smooth muscle cell migration(GO:1904754) |
| 0.0 | 1.9 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.8 | GO:0030163 | protein catabolic process(GO:0030163) |
| 0.0 | 2.7 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.3 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 1.3 | GO:0003170 | heart valve development(GO:0003170) |
| 0.0 | 0.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 1.8 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 6.1 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 0.8 | GO:0015991 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 3.1 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 2.9 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 0.3 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.8 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 2.7 | GO:0000398 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 9.2 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 1.4 | 4.3 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 1.3 | 5.0 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 1.0 | 2.9 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.9 | 3.7 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.4 | 3.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.3 | 1.5 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.2 | 2.9 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.2 | 2.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 3.2 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 2.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 4.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 1.2 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.1 | 2.7 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 1.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 0.8 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 3.2 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 2.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 3.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.8 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.7 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.9 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.6 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 1.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.7 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 2.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 1.2 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 11.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 3.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 4.7 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 5.6 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 5.3 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 3.3 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 4.4 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0005118 | sevenless binding(GO:0005118) |
| 1.6 | 6.3 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 1.3 | 3.9 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 1.2 | 4.7 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.8 | 4.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.8 | 6.4 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.8 | 2.3 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.7 | 2.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.6 | 12.3 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.4 | 3.2 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.3 | 5.0 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.3 | 3.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.3 | 5.0 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.2 | 1.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.2 | 2.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 2.9 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 0.2 | 3.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 0.9 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.2 | 3.5 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 4.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 5.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 2.7 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.7 | GO:0001093 | TFIIB-class transcription factor binding(GO:0001093) |
| 0.1 | 1.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 1.7 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 2.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 12.2 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 1.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 2.9 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.4 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 3.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 2.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 2.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.8 | GO:0043747 | protein-N-terminal asparagine amidohydrolase activity(GO:0008418) UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase activity(GO:0008759) iprodione amidohydrolase activity(GO:0018748) (3,5-dichlorophenylurea)acetate amidohydrolase activity(GO:0018749) 4'-(2-hydroxyisopropyl)phenylurea amidohydrolase activity(GO:0034571) didemethylisoproturon amidohydrolase activity(GO:0034573) N-isopropylacetanilide amidohydrolase activity(GO:0034576) N-cyclohexylformamide amidohydrolase activity(GO:0034781) isonicotinic acid hydrazide hydrolase activity(GO:0034876) cis-aconitamide amidase activity(GO:0034882) gamma-N-formylaminovinylacetate hydrolase activity(GO:0034885) N2-acetyl-L-lysine deacetylase activity(GO:0043747) O-succinylbenzoate synthase activity(GO:0043748) indoleacetamide hydrolase activity(GO:0043864) N-acetylcitrulline deacetylase activity(GO:0043909) N-acetylgalactosamine-6-phosphate deacetylase activity(GO:0047419) diacetylchitobiose deacetylase activity(GO:0052773) chitooligosaccharide deacetylase activity(GO:0052790) |
| 0.0 | 4.4 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 6.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 7.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.7 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 3.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 4.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.6 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 4.4 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 0.6 | GO:0048365 | Rac GTPase binding(GO:0048365) |