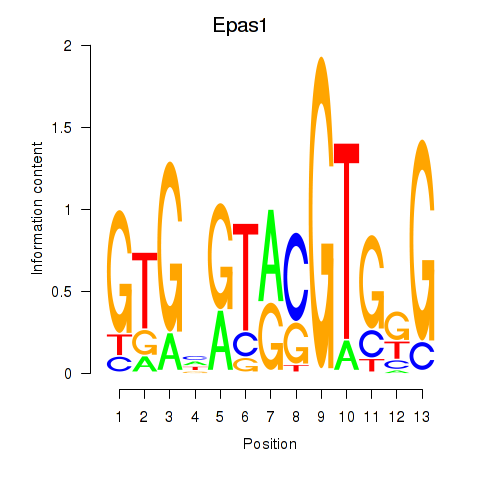
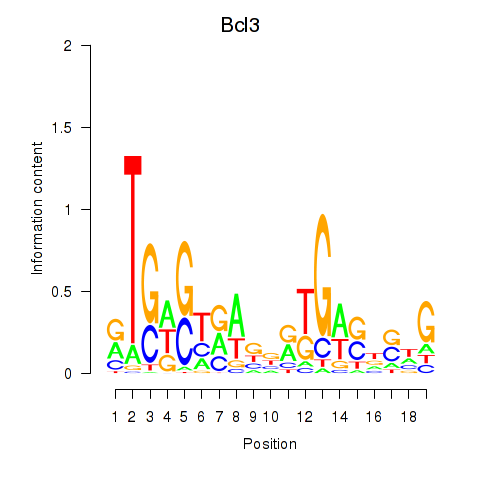
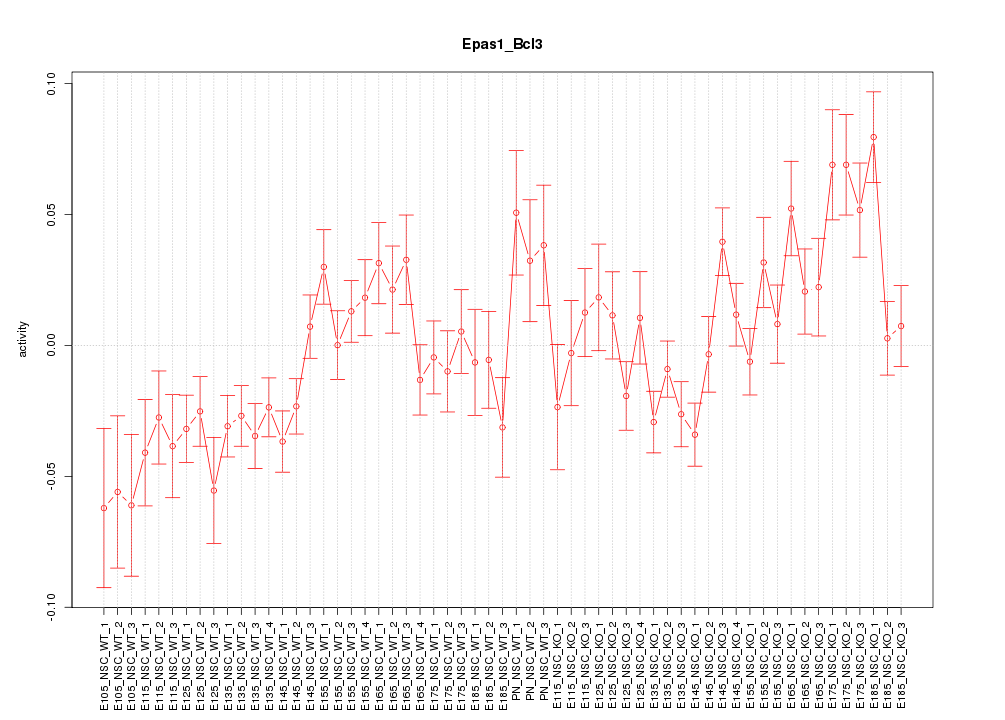
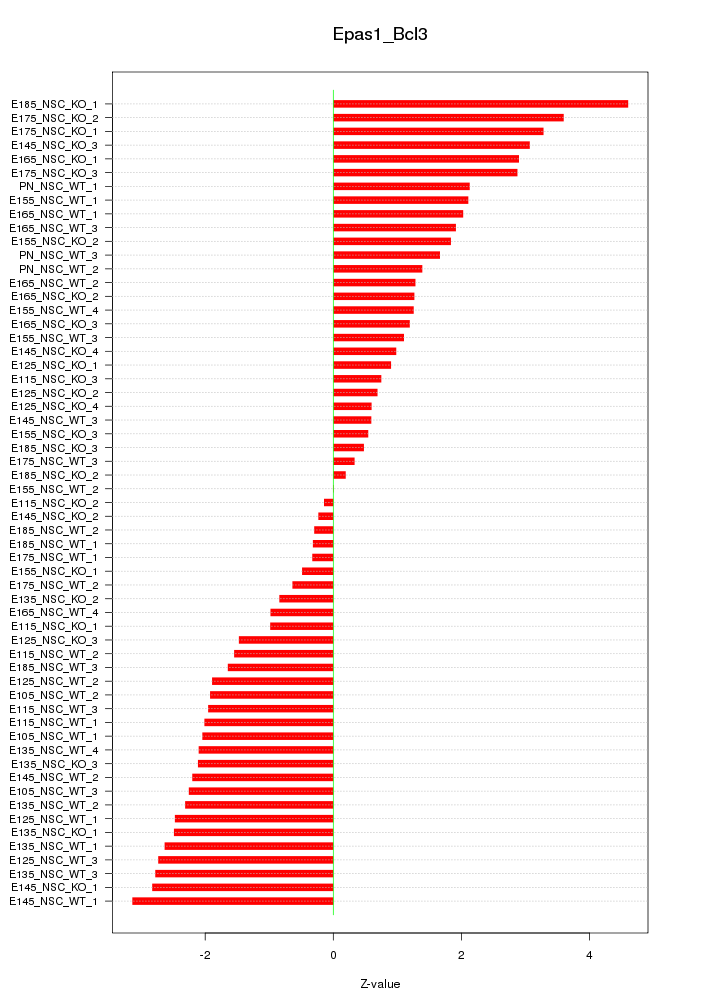
| 6.2 |
18.7 |
GO:0038095 |
positive regulation of mast cell cytokine production(GO:0032765) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 4.0 |
23.8 |
GO:0007217 |
tachykinin receptor signaling pathway(GO:0007217) |
| 3.8 |
26.9 |
GO:0061092 |
regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 3.4 |
10.2 |
GO:0050915 |
sensory perception of sour taste(GO:0050915) |
| 3.3 |
13.2 |
GO:0051562 |
negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 2.7 |
16.0 |
GO:0046880 |
regulation of follicle-stimulating hormone secretion(GO:0046880) follicle-stimulating hormone secretion(GO:0046884) |
| 2.3 |
6.8 |
GO:0098528 |
terpenoid catabolic process(GO:0016115) skeletal muscle fiber differentiation(GO:0098528) |
| 2.3 |
11.3 |
GO:0042699 |
follicle-stimulating hormone signaling pathway(GO:0042699) |
| 2.0 |
11.8 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 1.9 |
7.5 |
GO:0055095 |
lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 1.8 |
10.9 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 1.8 |
14.3 |
GO:0097264 |
self proteolysis(GO:0097264) |
| 1.6 |
4.7 |
GO:0007621 |
negative regulation of female receptivity(GO:0007621) |
| 1.5 |
9.2 |
GO:1902866 |
regulation of retina development in camera-type eye(GO:1902866) |
| 1.4 |
6.8 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 1.3 |
4.0 |
GO:1903795 |
regulation of inorganic anion transmembrane transport(GO:1903795) |
| 1.3 |
2.6 |
GO:0001803 |
type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 1.3 |
3.9 |
GO:1900736 |
regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 1.2 |
6.2 |
GO:0031133 |
regulation of axon diameter(GO:0031133) |
| 1.2 |
10.8 |
GO:0036371 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 1.2 |
5.9 |
GO:0075136 |
response to defenses of other organism involved in symbiotic interaction(GO:0052173) response to host defenses(GO:0052200) response to host(GO:0075136) |
| 1.2 |
5.8 |
GO:0050908 |
detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 1.1 |
10.7 |
GO:0071435 |
potassium ion export(GO:0071435) |
| 1.0 |
3.1 |
GO:1901228 |
regulation of osteoclast proliferation(GO:0090289) negative regulation of bone mineralization involved in bone maturation(GO:1900158) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 1.0 |
3.9 |
GO:0032227 |
negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 1.0 |
2.9 |
GO:0048686 |
regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.9 |
7.5 |
GO:0001542 |
ovulation from ovarian follicle(GO:0001542) |
| 0.9 |
4.5 |
GO:0034436 |
glycoprotein transport(GO:0034436) |
| 0.9 |
13.3 |
GO:0051481 |
negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.9 |
14.1 |
GO:0014051 |
gamma-aminobutyric acid secretion(GO:0014051) |
| 0.9 |
2.6 |
GO:1901731 |
calcium-mediated signaling using extracellular calcium source(GO:0035585) positive regulation of platelet aggregation(GO:1901731) |
| 0.9 |
2.6 |
GO:0002447 |
eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.8 |
4.2 |
GO:0018125 |
peptidyl-cysteine methylation(GO:0018125) |
| 0.8 |
3.4 |
GO:0097167 |
circadian regulation of translation(GO:0097167) |
| 0.8 |
8.0 |
GO:0090232 |
positive regulation of spindle checkpoint(GO:0090232) |
| 0.8 |
16.7 |
GO:0035235 |
ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.8 |
6.1 |
GO:0032264 |
IMP salvage(GO:0032264) |
| 0.7 |
5.2 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.7 |
1.5 |
GO:0002158 |
osteoclast proliferation(GO:0002158) |
| 0.7 |
4.3 |
GO:0089711 |
D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) L-glutamate transmembrane transport(GO:0089711) |
| 0.7 |
4.3 |
GO:0033600 |
negative regulation of mammary gland epithelial cell proliferation(GO:0033600) regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.7 |
2.1 |
GO:0006030 |
chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.7 |
4.2 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.7 |
3.4 |
GO:0010724 |
regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.7 |
2.0 |
GO:0031086 |
nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.6 |
3.9 |
GO:2000807 |
regulation of synaptic vesicle clustering(GO:2000807) |
| 0.6 |
4.3 |
GO:0045650 |
negative regulation of macrophage differentiation(GO:0045650) |
| 0.6 |
1.7 |
GO:0034162 |
toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.5 |
7.1 |
GO:0043252 |
prostaglandin transport(GO:0015732) sodium-independent organic anion transport(GO:0043252) |
| 0.5 |
1.1 |
GO:0010958 |
regulation of amino acid import(GO:0010958) regulation of L-arginine import(GO:0010963) |
| 0.5 |
6.5 |
GO:0035988 |
chondrocyte proliferation(GO:0035988) |
| 0.5 |
3.8 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.5 |
3.2 |
GO:0061343 |
cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.5 |
1.6 |
GO:0003289 |
atrial septum primum morphogenesis(GO:0003289) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.5 |
10.1 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.5 |
6.5 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.5 |
1.9 |
GO:0035426 |
extracellular matrix-cell signaling(GO:0035426) |
| 0.5 |
2.3 |
GO:0001778 |
plasma membrane repair(GO:0001778) |
| 0.5 |
9.2 |
GO:0050650 |
chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.4 |
2.6 |
GO:0021631 |
optic nerve morphogenesis(GO:0021631) |
| 0.4 |
1.8 |
GO:0006049 |
UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.4 |
1.6 |
GO:0035521 |
monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.4 |
10.8 |
GO:0010738 |
regulation of protein kinase A signaling(GO:0010738) |
| 0.4 |
17.1 |
GO:1900449 |
regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.4 |
1.2 |
GO:1902277 |
negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.4 |
2.7 |
GO:0033563 |
dorsal/ventral axon guidance(GO:0033563) |
| 0.4 |
1.2 |
GO:0002337 |
B-1a B cell differentiation(GO:0002337) patterning of lymph vessels(GO:0060854) |
| 0.4 |
1.2 |
GO:0030070 |
insulin processing(GO:0030070) |
| 0.4 |
3.3 |
GO:0031547 |
brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.4 |
1.8 |
GO:0043314 |
negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.4 |
6.0 |
GO:0099500 |
synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.3 |
1.0 |
GO:0002513 |
tolerance induction to self antigen(GO:0002513) |
| 0.3 |
1.7 |
GO:0032423 |
regulation of mismatch repair(GO:0032423) |
| 0.3 |
3.3 |
GO:0036159 |
inner dynein arm assembly(GO:0036159) |
| 0.3 |
6.7 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.3 |
8.2 |
GO:1901381 |
positive regulation of potassium ion transmembrane transport(GO:1901381) |
| 0.3 |
1.4 |
GO:0021856 |
cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.3 |
2.9 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.3 |
9.9 |
GO:0051930 |
regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.3 |
1.8 |
GO:1903025 |
regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.2 |
2.2 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.2 |
9.8 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.2 |
0.9 |
GO:0043415 |
positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.2 |
1.7 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.2 |
21.6 |
GO:0002028 |
regulation of sodium ion transport(GO:0002028) |
| 0.2 |
1.3 |
GO:0048170 |
positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.2 |
0.7 |
GO:0070649 |
formin-nucleated actin cable assembly(GO:0070649) |
| 0.2 |
3.7 |
GO:1903861 |
regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.2 |
2.3 |
GO:0016082 |
synaptic vesicle priming(GO:0016082) |
| 0.2 |
3.0 |
GO:0045599 |
negative regulation of fat cell differentiation(GO:0045599) |
| 0.2 |
1.0 |
GO:0070493 |
thrombin receptor signaling pathway(GO:0070493) |
| 0.2 |
0.4 |
GO:1900126 |
negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.2 |
3.1 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.2 |
3.9 |
GO:0035725 |
sodium ion transmembrane transport(GO:0035725) |
| 0.2 |
2.6 |
GO:0006622 |
protein targeting to lysosome(GO:0006622) |
| 0.2 |
5.8 |
GO:0060074 |
synapse maturation(GO:0060074) |
| 0.2 |
5.7 |
GO:0008045 |
motor neuron axon guidance(GO:0008045) |
| 0.2 |
0.5 |
GO:1902310 |
positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.2 |
1.3 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.2 |
6.1 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.2 |
3.1 |
GO:0061029 |
eyelid development in camera-type eye(GO:0061029) |
| 0.2 |
3.3 |
GO:0048026 |
positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 |
1.2 |
GO:0006265 |
DNA topological change(GO:0006265) |
| 0.1 |
0.6 |
GO:0043461 |
proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.1 |
1.8 |
GO:0033623 |
regulation of integrin activation(GO:0033623) |
| 0.1 |
0.4 |
GO:0071879 |
positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.1 |
3.9 |
GO:0061098 |
positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.1 |
5.0 |
GO:0097178 |
ruffle assembly(GO:0097178) |
| 0.1 |
1.3 |
GO:0021869 |
forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 |
9.0 |
GO:0001578 |
microtubule bundle formation(GO:0001578) |
| 0.1 |
0.2 |
GO:1905005 |
regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905005) |
| 0.1 |
10.2 |
GO:0043149 |
contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.1 |
7.7 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.1 |
0.3 |
GO:0034729 |
histone H3-K79 methylation(GO:0034729) |
| 0.1 |
3.1 |
GO:0007189 |
adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.1 |
2.1 |
GO:0003073 |
regulation of systemic arterial blood pressure(GO:0003073) |
| 0.1 |
2.4 |
GO:0034340 |
response to type I interferon(GO:0034340) |
| 0.1 |
1.4 |
GO:0043508 |
negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 |
5.9 |
GO:0006749 |
glutathione metabolic process(GO:0006749) |
| 0.1 |
3.6 |
GO:0008542 |
visual learning(GO:0008542) |
| 0.1 |
1.3 |
GO:0050765 |
negative regulation of phagocytosis(GO:0050765) |
| 0.1 |
3.1 |
GO:0071277 |
cellular response to calcium ion(GO:0071277) |
| 0.1 |
0.3 |
GO:0031069 |
hair follicle morphogenesis(GO:0031069) |
| 0.1 |
12.7 |
GO:0010977 |
negative regulation of neuron projection development(GO:0010977) |
| 0.1 |
0.2 |
GO:1900623 |
regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 |
8.4 |
GO:0006165 |
nucleoside diphosphate phosphorylation(GO:0006165) |
| 0.1 |
0.4 |
GO:0045605 |
negative regulation of epidermal cell differentiation(GO:0045605) |
| 0.1 |
1.1 |
GO:0070935 |
3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 |
1.8 |
GO:0045747 |
positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 |
2.3 |
GO:0000381 |
regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 |
4.3 |
GO:0030968 |
endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 |
0.2 |
GO:0006682 |
galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 |
1.0 |
GO:0006044 |
N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 |
2.9 |
GO:0051298 |
centrosome duplication(GO:0051298) |
| 0.0 |
0.3 |
GO:0035372 |
protein localization to microtubule(GO:0035372) |
| 0.0 |
6.9 |
GO:0051291 |
protein heterooligomerization(GO:0051291) |
| 0.0 |
0.4 |
GO:1902902 |
negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 |
0.8 |
GO:0035036 |
binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.0 |
1.8 |
GO:0046513 |
ceramide biosynthetic process(GO:0046513) |
| 0.0 |
2.2 |
GO:0032091 |
negative regulation of protein binding(GO:0032091) |
| 0.0 |
0.2 |
GO:2000483 |
negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 |
1.1 |
GO:0000266 |
mitochondrial fission(GO:0000266) |
| 0.0 |
2.1 |
GO:0051965 |
positive regulation of synapse assembly(GO:0051965) |
| 0.0 |
2.9 |
GO:0098792 |
xenophagy(GO:0098792) |
| 0.0 |
1.2 |
GO:0006986 |
response to unfolded protein(GO:0006986) |
| 0.0 |
1.9 |
GO:0043484 |
regulation of RNA splicing(GO:0043484) |
| 0.0 |
2.5 |
GO:0048515 |
spermatid differentiation(GO:0048515) |
| 0.0 |
0.4 |
GO:0032402 |
melanosome transport(GO:0032402) |
| 0.0 |
1.0 |
GO:0007032 |
endosome organization(GO:0007032) |
| 0.0 |
1.5 |
GO:0006402 |
mRNA catabolic process(GO:0006402) |
| 0.0 |
0.3 |
GO:0042059 |
negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.0 |
0.9 |
GO:0006754 |
ATP biosynthetic process(GO:0006754) |
| 0.0 |
0.1 |
GO:0019236 |
response to pheromone(GO:0019236) |
| 0.0 |
1.3 |
GO:0030183 |
B cell differentiation(GO:0030183) |
| 0.0 |
2.1 |
GO:0007623 |
circadian rhythm(GO:0007623) |
| 0.0 |
0.4 |
GO:0010811 |
positive regulation of cell-substrate adhesion(GO:0010811) |