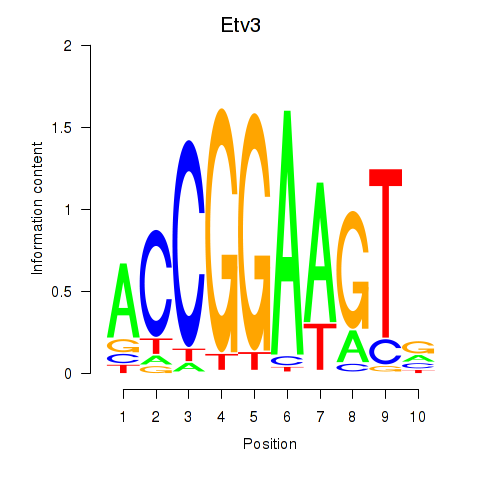
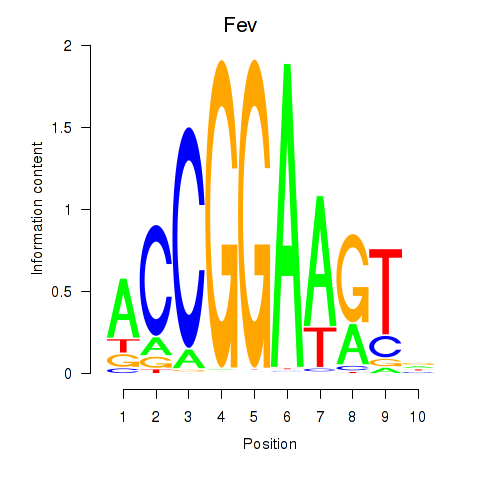
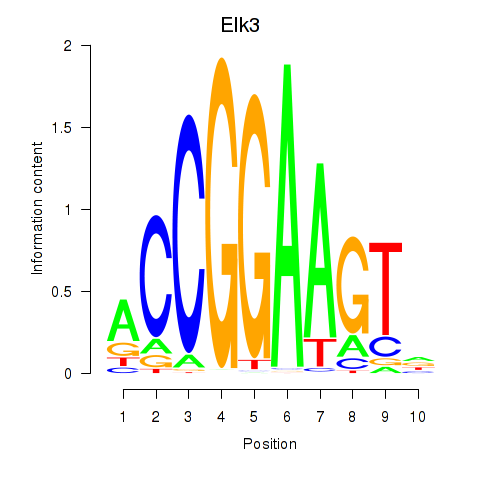
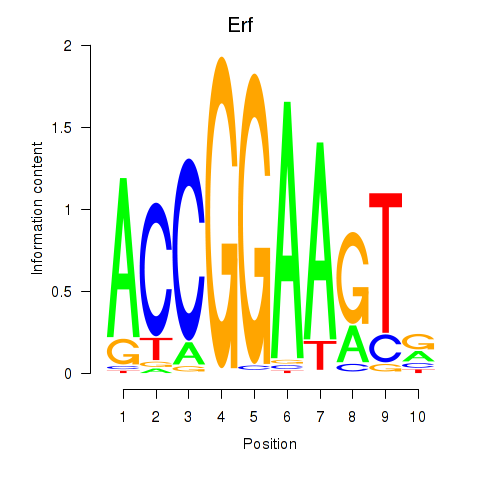
| 4.1 |
12.2 |
GO:0048611 |
ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 3.0 |
24.0 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 2.6 |
7.8 |
GO:0019085 |
early viral transcription(GO:0019085) |
| 2.6 |
5.2 |
GO:1901668 |
regulation of superoxide dismutase activity(GO:1901668) |
| 2.0 |
7.8 |
GO:0018307 |
enzyme active site formation(GO:0018307) |
| 1.9 |
5.8 |
GO:0071929 |
alpha-tubulin acetylation(GO:0071929) |
| 1.6 |
4.8 |
GO:0042998 |
positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 1.5 |
4.6 |
GO:1902528 |
regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 1.5 |
5.9 |
GO:0086042 |
cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) |
| 1.4 |
5.7 |
GO:0055071 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 1.3 |
5.3 |
GO:0044565 |
dendritic cell proliferation(GO:0044565) |
| 1.3 |
3.8 |
GO:0051542 |
elastin biosynthetic process(GO:0051542) |
| 1.2 |
3.5 |
GO:1901254 |
positive regulation of intracellular transport of viral material(GO:1901254) |
| 1.1 |
3.3 |
GO:0070124 |
mitochondrial translational initiation(GO:0070124) |
| 1.1 |
5.4 |
GO:0021764 |
amygdala development(GO:0021764) |
| 1.1 |
3.2 |
GO:0036092 |
phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 1.0 |
3.1 |
GO:0043323 |
positive regulation of natural killer cell degranulation(GO:0043323) |
| 1.0 |
11.3 |
GO:0051103 |
DNA ligation involved in DNA repair(GO:0051103) |
| 1.0 |
4.1 |
GO:0007228 |
positive regulation of hh target transcription factor activity(GO:0007228) |
| 1.0 |
3.9 |
GO:0070127 |
tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 1.0 |
4.8 |
GO:0030242 |
pexophagy(GO:0030242) |
| 0.9 |
2.8 |
GO:0090187 |
positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.9 |
5.6 |
GO:1903690 |
negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.9 |
15.8 |
GO:0045792 |
negative regulation of cell size(GO:0045792) |
| 0.9 |
5.4 |
GO:0044034 |
negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.9 |
2.6 |
GO:0043201 |
response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.8 |
4.9 |
GO:0006266 |
DNA ligation(GO:0006266) |
| 0.8 |
4.0 |
GO:2000189 |
positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.8 |
5.5 |
GO:1904217 |
regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.8 |
3.1 |
GO:0090365 |
regulation of mRNA modification(GO:0090365) |
| 0.8 |
12.2 |
GO:0060363 |
cranial suture morphogenesis(GO:0060363) |
| 0.7 |
3.0 |
GO:2000812 |
regulation of barbed-end actin filament capping(GO:2000812) |
| 0.7 |
2.2 |
GO:0002331 |
pre-B cell allelic exclusion(GO:0002331) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.7 |
2.2 |
GO:0051342 |
regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.7 |
2.2 |
GO:0018103 |
protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.7 |
2.9 |
GO:0046898 |
response to cycloheximide(GO:0046898) |
| 0.7 |
6.3 |
GO:1901725 |
regulation of histone deacetylase activity(GO:1901725) |
| 0.7 |
1.4 |
GO:0070889 |
platelet alpha granule organization(GO:0070889) |
| 0.7 |
2.1 |
GO:0016191 |
synaptic vesicle uncoating(GO:0016191) regulation of endosome organization(GO:1904978) positive regulation of endosome organization(GO:1904980) |
| 0.7 |
2.1 |
GO:0034729 |
histone H3-K79 methylation(GO:0034729) |
| 0.7 |
2.8 |
GO:2000211 |
regulation of glutamate metabolic process(GO:2000211) |
| 0.7 |
2.0 |
GO:0060785 |
regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.7 |
2.6 |
GO:0060715 |
syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.6 |
2.6 |
GO:0097039 |
protein linear polyubiquitination(GO:0097039) |
| 0.6 |
6.4 |
GO:0043517 |
positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.6 |
3.1 |
GO:2000574 |
regulation of microtubule motor activity(GO:2000574) |
| 0.6 |
1.8 |
GO:2000834 |
androgen secretion(GO:0035935) testosterone secretion(GO:0035936) regulation of androgen secretion(GO:2000834) positive regulation of androgen secretion(GO:2000836) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.6 |
6.6 |
GO:0030157 |
pancreatic juice secretion(GO:0030157) |
| 0.6 |
2.4 |
GO:0071105 |
response to interleukin-11(GO:0071105) osteoclast fusion(GO:0072675) |
| 0.6 |
2.3 |
GO:0035617 |
stress granule disassembly(GO:0035617) |
| 0.6 |
3.9 |
GO:0071712 |
ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.5 |
1.6 |
GO:0003245 |
cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.5 |
2.2 |
GO:0002069 |
columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.5 |
10.7 |
GO:0030033 |
microvillus assembly(GO:0030033) |
| 0.5 |
2.7 |
GO:0071894 |
histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.5 |
2.1 |
GO:0034454 |
microtubule anchoring at centrosome(GO:0034454) |
| 0.5 |
1.6 |
GO:1903903 |
regulation of establishment of T cell polarity(GO:1903903) |
| 0.5 |
1.1 |
GO:0061511 |
centriole elongation(GO:0061511) |
| 0.5 |
1.6 |
GO:0014042 |
positive regulation of mitochondrial fusion(GO:0010636) positive regulation of neuron maturation(GO:0014042) |
| 0.5 |
1.5 |
GO:0036500 |
ATF6-mediated unfolded protein response(GO:0036500) |
| 0.5 |
3.0 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.5 |
3.0 |
GO:0043983 |
histone H4-K12 acetylation(GO:0043983) |
| 0.5 |
1.4 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.5 |
2.4 |
GO:0006680 |
glucosylceramide catabolic process(GO:0006680) |
| 0.5 |
2.9 |
GO:0001302 |
replicative cell aging(GO:0001302) |
| 0.5 |
1.9 |
GO:0034975 |
protein folding in endoplasmic reticulum(GO:0034975) |
| 0.5 |
4.3 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.5 |
2.3 |
GO:0060050 |
positive regulation of protein glycosylation(GO:0060050) |
| 0.5 |
3.7 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.4 |
0.4 |
GO:0055005 |
ventricular cardiac myofibril assembly(GO:0055005) |
| 0.4 |
9.4 |
GO:0000301 |
retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.4 |
2.7 |
GO:0070863 |
positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.4 |
2.6 |
GO:0098789 |
pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.4 |
3.0 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.4 |
2.1 |
GO:0015871 |
choline transport(GO:0015871) |
| 0.4 |
0.8 |
GO:0090611 |
ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.4 |
2.5 |
GO:0048102 |
autophagic cell death(GO:0048102) |
| 0.4 |
3.7 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.4 |
4.4 |
GO:0034497 |
protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.4 |
1.2 |
GO:0016344 |
meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.4 |
1.2 |
GO:1903774 |
positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.4 |
1.2 |
GO:0090327 |
negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.4 |
1.9 |
GO:0034058 |
endosomal vesicle fusion(GO:0034058) |
| 0.4 |
2.3 |
GO:0016557 |
peroxisome membrane biogenesis(GO:0016557) |
| 0.4 |
1.2 |
GO:0045014 |
carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.4 |
1.1 |
GO:0060800 |
regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.4 |
8.7 |
GO:0002076 |
osteoblast development(GO:0002076) |
| 0.4 |
1.5 |
GO:0006049 |
UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.4 |
6.0 |
GO:2000821 |
AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) regulation of grooming behavior(GO:2000821) |
| 0.4 |
1.9 |
GO:0036258 |
multivesicular body assembly(GO:0036258) |
| 0.4 |
1.5 |
GO:0009212 |
dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.4 |
3.3 |
GO:0071569 |
protein ufmylation(GO:0071569) |
| 0.4 |
2.6 |
GO:0006102 |
isocitrate metabolic process(GO:0006102) |
| 0.4 |
2.9 |
GO:0031848 |
protection from non-homologous end joining at telomere(GO:0031848) |
| 0.4 |
2.9 |
GO:0019348 |
dolichol metabolic process(GO:0019348) |
| 0.4 |
6.3 |
GO:2000369 |
regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.3 |
0.7 |
GO:0042726 |
flavin-containing compound metabolic process(GO:0042726) |
| 0.3 |
1.4 |
GO:0061010 |
gall bladder development(GO:0061010) |
| 0.3 |
14.7 |
GO:0036465 |
synaptic vesicle recycling(GO:0036465) |
| 0.3 |
1.7 |
GO:1903071 |
positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.3 |
5.1 |
GO:0070816 |
phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.3 |
1.0 |
GO:0097155 |
fasciculation of sensory neuron axon(GO:0097155) |
| 0.3 |
2.3 |
GO:2000643 |
positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.3 |
1.6 |
GO:0006543 |
glutamine catabolic process(GO:0006543) |
| 0.3 |
2.3 |
GO:0050882 |
voluntary musculoskeletal movement(GO:0050882) |
| 0.3 |
1.0 |
GO:0046881 |
sperm ejaculation(GO:0042713) positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.3 |
2.0 |
GO:0060046 |
regulation of acrosome reaction(GO:0060046) |
| 0.3 |
1.9 |
GO:0070072 |
vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.3 |
1.3 |
GO:0051771 |
negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.3 |
4.1 |
GO:0003283 |
atrial septum development(GO:0003283) |
| 0.3 |
1.7 |
GO:0006123 |
mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.3 |
2.0 |
GO:0006361 |
transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.3 |
2.9 |
GO:1904152 |
negative regulation of protein exit from endoplasmic reticulum(GO:0070862) regulation of retrograde protein transport, ER to cytosol(GO:1904152) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.3 |
0.9 |
GO:0051031 |
tRNA transport(GO:0051031) |
| 0.3 |
1.1 |
GO:0006481 |
C-terminal protein methylation(GO:0006481) |
| 0.3 |
1.4 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.3 |
1.1 |
GO:1900017 |
positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.3 |
2.2 |
GO:0000395 |
mRNA 5'-splice site recognition(GO:0000395) |
| 0.3 |
1.1 |
GO:2000049 |
positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.3 |
3.0 |
GO:0097151 |
positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.3 |
0.3 |
GO:0002034 |
angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.3 |
1.9 |
GO:0006701 |
progesterone biosynthetic process(GO:0006701) |
| 0.3 |
1.3 |
GO:1900095 |
regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.3 |
8.2 |
GO:0006829 |
zinc II ion transport(GO:0006829) |
| 0.3 |
0.8 |
GO:0036508 |
protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) protein demannosylation(GO:0036507) protein alpha-1,2-demannosylation(GO:0036508) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.2 |
2.2 |
GO:2000650 |
negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.2 |
2.5 |
GO:1901621 |
negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.2 |
2.7 |
GO:0035646 |
endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.2 |
1.0 |
GO:0010847 |
regulation of chromatin assembly(GO:0010847) |
| 0.2 |
2.4 |
GO:0031665 |
negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.2 |
0.7 |
GO:0006030 |
chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.2 |
1.4 |
GO:0071639 |
positive regulation of monocyte chemotactic protein-1 production(GO:0071639) |
| 0.2 |
0.7 |
GO:0097500 |
receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.2 |
2.5 |
GO:0006622 |
protein targeting to lysosome(GO:0006622) |
| 0.2 |
2.7 |
GO:0007250 |
activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.2 |
6.5 |
GO:0015991 |
energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.2 |
2.0 |
GO:0071044 |
histone mRNA catabolic process(GO:0071044) |
| 0.2 |
2.6 |
GO:2000096 |
positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.2 |
1.7 |
GO:0030836 |
positive regulation of actin filament depolymerization(GO:0030836) |
| 0.2 |
9.6 |
GO:0048278 |
vesicle docking(GO:0048278) |
| 0.2 |
2.1 |
GO:0044804 |
nucleophagy(GO:0044804) |
| 0.2 |
0.6 |
GO:0090529 |
barrier septum assembly(GO:0000917) cell septum assembly(GO:0090529) |
| 0.2 |
0.6 |
GO:0010635 |
regulation of mitochondrial fusion(GO:0010635) |
| 0.2 |
9.2 |
GO:0045761 |
regulation of adenylate cyclase activity(GO:0045761) |
| 0.2 |
0.6 |
GO:1902071 |
positive regulation of cellular response to hypoxia(GO:1900039) regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.2 |
1.5 |
GO:0038031 |
non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.2 |
2.5 |
GO:1904424 |
regulation of GTP binding(GO:1904424) |
| 0.2 |
13.2 |
GO:0003281 |
ventricular septum development(GO:0003281) |
| 0.2 |
1.8 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.2 |
0.4 |
GO:1904431 |
positive regulation of t-circle formation(GO:1904431) |
| 0.2 |
2.0 |
GO:0051533 |
positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.2 |
0.4 |
GO:0000350 |
generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.2 |
2.0 |
GO:1990440 |
positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.2 |
0.6 |
GO:0021691 |
cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.2 |
1.3 |
GO:1903895 |
negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 0.2 |
0.6 |
GO:0031117 |
positive regulation of microtubule depolymerization(GO:0031117) |
| 0.2 |
3.1 |
GO:0072583 |
clathrin-mediated endocytosis(GO:0072583) |
| 0.2 |
2.9 |
GO:0016973 |
poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.2 |
1.6 |
GO:0070995 |
NADPH oxidation(GO:0070995) |
| 0.2 |
2.7 |
GO:0006750 |
glutathione biosynthetic process(GO:0006750) |
| 0.2 |
0.5 |
GO:0001828 |
inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.2 |
0.5 |
GO:0031642 |
negative regulation of myelination(GO:0031642) |
| 0.2 |
1.4 |
GO:1900037 |
regulation of cellular response to hypoxia(GO:1900037) |
| 0.2 |
4.2 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 |
1.2 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.2 |
0.9 |
GO:0045144 |
meiotic sister chromatid segregation(GO:0045144) meiotic sister chromatid cohesion(GO:0051177) |
| 0.2 |
0.5 |
GO:0071677 |
positive regulation of mononuclear cell migration(GO:0071677) |
| 0.2 |
0.2 |
GO:0043313 |
regulation of neutrophil degranulation(GO:0043313) |
| 0.2 |
0.5 |
GO:0032241 |
positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.2 |
0.5 |
GO:0010626 |
negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.2 |
1.3 |
GO:0048280 |
vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.2 |
1.5 |
GO:0021554 |
optic nerve development(GO:0021554) |
| 0.2 |
0.7 |
GO:0031937 |
positive regulation of chromatin silencing(GO:0031937) |
| 0.2 |
0.3 |
GO:0010724 |
regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.2 |
0.7 |
GO:1901524 |
regulation of macromitophagy(GO:1901524) negative regulation of macromitophagy(GO:1901525) |
| 0.2 |
0.5 |
GO:1903715 |
regulation of aerobic respiration(GO:1903715) |
| 0.2 |
0.8 |
GO:1902897 |
regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.2 |
3.3 |
GO:0051457 |
maintenance of protein location in nucleus(GO:0051457) |
| 0.2 |
0.6 |
GO:0071139 |
resolution of recombination intermediates(GO:0071139) |
| 0.2 |
1.2 |
GO:0046007 |
negative regulation of activated T cell proliferation(GO:0046007) |
| 0.2 |
0.5 |
GO:0036228 |
protein targeting to nuclear inner membrane(GO:0036228) |
| 0.1 |
1.8 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.1 |
0.9 |
GO:0006995 |
cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 |
0.4 |
GO:0070535 |
histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.1 |
0.1 |
GO:0048227 |
plasma membrane to endosome transport(GO:0048227) |
| 0.1 |
0.7 |
GO:0098543 |
detection of bacterium(GO:0016045) detection of other organism(GO:0098543) |
| 0.1 |
2.7 |
GO:0071800 |
podosome assembly(GO:0071800) |
| 0.1 |
0.6 |
GO:0035752 |
lysosomal lumen pH elevation(GO:0035752) |
| 0.1 |
0.7 |
GO:0045347 |
negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 |
0.8 |
GO:0034983 |
peptidyl-lysine deacetylation(GO:0034983) |
| 0.1 |
1.0 |
GO:0099515 |
actin filament-based transport(GO:0099515) |
| 0.1 |
1.1 |
GO:2000601 |
positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 |
0.7 |
GO:0006983 |
ER overload response(GO:0006983) |
| 0.1 |
4.9 |
GO:0045668 |
negative regulation of osteoblast differentiation(GO:0045668) |
| 0.1 |
0.5 |
GO:0030397 |
membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.1 |
4.0 |
GO:0007041 |
lysosomal transport(GO:0007041) |
| 0.1 |
1.4 |
GO:0006895 |
Golgi to endosome transport(GO:0006895) |
| 0.1 |
1.0 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.1 |
0.4 |
GO:0008612 |
peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.1 |
0.6 |
GO:0045213 |
neurotransmitter receptor metabolic process(GO:0045213) |
| 0.1 |
2.2 |
GO:0044550 |
secondary metabolite biosynthetic process(GO:0044550) |
| 0.1 |
0.9 |
GO:0060294 |
cilium movement involved in cell motility(GO:0060294) |
| 0.1 |
0.6 |
GO:0070966 |
nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 |
1.0 |
GO:1901844 |
regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.1 |
0.1 |
GO:1903599 |
positive regulation of mitophagy(GO:1903599) |
| 0.1 |
5.1 |
GO:0051865 |
protein autoubiquitination(GO:0051865) |
| 0.1 |
1.0 |
GO:0007175 |
negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 |
2.2 |
GO:2000311 |
regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 |
0.6 |
GO:0034227 |
tRNA thio-modification(GO:0034227) |
| 0.1 |
0.8 |
GO:0021942 |
radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 |
0.4 |
GO:0042275 |
error-free postreplication DNA repair(GO:0042275) |
| 0.1 |
4.9 |
GO:0007257 |
activation of JUN kinase activity(GO:0007257) |
| 0.1 |
0.5 |
GO:0006544 |
glycine metabolic process(GO:0006544) |
| 0.1 |
0.3 |
GO:0097343 |
ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.1 |
1.0 |
GO:0001672 |
regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 |
2.6 |
GO:0034314 |
Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 |
1.1 |
GO:0051601 |
exocyst localization(GO:0051601) |
| 0.1 |
0.7 |
GO:0001539 |
cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.1 |
2.1 |
GO:0043507 |
positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 |
1.1 |
GO:0006515 |
misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.1 |
0.6 |
GO:0061339 |
establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.1 |
3.5 |
GO:0061512 |
protein localization to cilium(GO:0061512) |
| 0.1 |
4.0 |
GO:0006513 |
protein monoubiquitination(GO:0006513) |
| 0.1 |
1.7 |
GO:0032967 |
positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.1 |
2.1 |
GO:0007032 |
endosome organization(GO:0007032) |
| 0.1 |
1.6 |
GO:0072673 |
lamellipodium morphogenesis(GO:0072673) |
| 0.1 |
0.4 |
GO:1902255 |
positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.1 |
0.2 |
GO:0051892 |
regulation of cardioblast differentiation(GO:0051890) negative regulation of cardioblast differentiation(GO:0051892) |
| 0.1 |
0.8 |
GO:2001256 |
regulation of store-operated calcium entry(GO:2001256) |
| 0.1 |
0.2 |
GO:0030950 |
establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 |
1.1 |
GO:0051001 |
negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.1 |
16.3 |
GO:0042787 |
protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.1 |
1.2 |
GO:1903799 |
negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.1 |
3.8 |
GO:0030521 |
androgen receptor signaling pathway(GO:0030521) |
| 0.1 |
6.1 |
GO:0035725 |
sodium ion transmembrane transport(GO:0035725) |
| 0.1 |
1.1 |
GO:0043649 |
dicarboxylic acid catabolic process(GO:0043649) |
| 0.1 |
1.0 |
GO:0045974 |
miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 |
0.5 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.1 |
1.3 |
GO:0035235 |
ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 |
0.4 |
GO:1901030 |
positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901030) |
| 0.1 |
0.6 |
GO:0099500 |
synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 |
0.7 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.1 |
0.3 |
GO:1903012 |
regulation of bone mineralization involved in bone maturation(GO:1900157) positive regulation of bone development(GO:1903012) |
| 0.1 |
0.4 |
GO:0010025 |
wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 |
0.4 |
GO:0015867 |
ATP transport(GO:0015867) |
| 0.1 |
2.6 |
GO:1902476 |
chloride transmembrane transport(GO:1902476) |
| 0.1 |
0.8 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 |
0.7 |
GO:0008105 |
asymmetric protein localization(GO:0008105) |
| 0.1 |
1.9 |
GO:0045197 |
establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.1 |
1.6 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.1 |
0.7 |
GO:0070932 |
histone H3 deacetylation(GO:0070932) |
| 0.1 |
0.2 |
GO:0070488 |
neutrophil aggregation(GO:0070488) |
| 0.1 |
0.3 |
GO:0030223 |
neutrophil differentiation(GO:0030223) |
| 0.1 |
0.6 |
GO:0090043 |
tubulin deacetylation(GO:0090042) regulation of tubulin deacetylation(GO:0090043) |
| 0.1 |
1.8 |
GO:2000036 |
regulation of stem cell population maintenance(GO:2000036) |
| 0.1 |
0.2 |
GO:0071985 |
multivesicular body sorting pathway(GO:0071985) |
| 0.1 |
0.1 |
GO:0042268 |
regulation of cytolysis(GO:0042268) |
| 0.1 |
0.2 |
GO:0090148 |
membrane fission(GO:0090148) |
| 0.1 |
0.3 |
GO:0043651 |
linoleic acid metabolic process(GO:0043651) |
| 0.1 |
3.0 |
GO:0042147 |
retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 |
1.4 |
GO:0045725 |
positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.1 |
0.1 |
GO:0010520 |
regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.1 |
1.1 |
GO:0015012 |
heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.1 |
0.6 |
GO:0051352 |
negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) |
| 0.1 |
0.7 |
GO:0030970 |
retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 |
0.6 |
GO:0006283 |
transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 |
2.1 |
GO:1901998 |
toxin transport(GO:1901998) |
| 0.1 |
2.9 |
GO:0031338 |
regulation of vesicle fusion(GO:0031338) |
| 0.1 |
0.6 |
GO:0060712 |
spongiotrophoblast layer development(GO:0060712) |
| 0.1 |
0.8 |
GO:0044110 |
growth involved in symbiotic interaction(GO:0044110) growth of symbiont involved in interaction with host(GO:0044116) growth of symbiont in host(GO:0044117) regulation of growth of symbiont in host(GO:0044126) modulation of growth of symbiont involved in interaction with host(GO:0044144) |
| 0.1 |
0.2 |
GO:0016561 |
protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.1 |
0.3 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.1 |
0.4 |
GO:1901837 |
negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.1 |
1.7 |
GO:0007212 |
dopamine receptor signaling pathway(GO:0007212) |
| 0.1 |
0.7 |
GO:1903020 |
positive regulation of glycoprotein metabolic process(GO:1903020) |
| 0.1 |
0.3 |
GO:0060059 |
embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.1 |
0.6 |
GO:0051683 |
establishment of Golgi localization(GO:0051683) |
| 0.1 |
0.2 |
GO:0006624 |
vacuolar protein processing(GO:0006624) |
| 0.1 |
0.5 |
GO:2000114 |
regulation of establishment of cell polarity(GO:2000114) |
| 0.1 |
0.1 |
GO:0045764 |
positive regulation of cellular amino acid metabolic process(GO:0045764) negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.1 |
1.0 |
GO:0007020 |
microtubule nucleation(GO:0007020) |
| 0.1 |
0.4 |
GO:0006370 |
7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 |
0.2 |
GO:0007000 |
nucleolus organization(GO:0007000) |
| 0.1 |
1.3 |
GO:0006890 |
retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 |
1.2 |
GO:0001706 |
endoderm formation(GO:0001706) |
| 0.1 |
8.3 |
GO:0010977 |
negative regulation of neuron projection development(GO:0010977) |
| 0.1 |
0.4 |
GO:0021756 |
striatum development(GO:0021756) |
| 0.1 |
1.6 |
GO:0007528 |
neuromuscular junction development(GO:0007528) |
| 0.1 |
0.2 |
GO:0042796 |
snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 |
0.3 |
GO:0046501 |
protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 |
3.4 |
GO:0050885 |
neuromuscular process controlling balance(GO:0050885) |
| 0.0 |
0.7 |
GO:0000731 |
DNA synthesis involved in DNA repair(GO:0000731) |
| 0.0 |
1.3 |
GO:0001678 |
cellular glucose homeostasis(GO:0001678) |
| 0.0 |
0.2 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 |
1.0 |
GO:0043330 |
response to exogenous dsRNA(GO:0043330) |
| 0.0 |
0.1 |
GO:0051030 |
snRNA transport(GO:0051030) |
| 0.0 |
1.6 |
GO:0050905 |
neuromuscular process(GO:0050905) |
| 0.0 |
1.8 |
GO:0008088 |
axo-dendritic transport(GO:0008088) |
| 0.0 |
0.3 |
GO:0080009 |
mRNA methylation(GO:0080009) |
| 0.0 |
2.8 |
GO:0035773 |
insulin secretion involved in cellular response to glucose stimulus(GO:0035773) |
| 0.0 |
0.3 |
GO:0042790 |
transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 |
0.2 |
GO:0040016 |
embryonic cleavage(GO:0040016) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 |
0.2 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 |
3.1 |
GO:0018107 |
peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 |
0.0 |
GO:0010936 |
negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 |
0.2 |
GO:1904892 |
regulation of JAK-STAT cascade(GO:0046425) regulation of STAT cascade(GO:1904892) |
| 0.0 |
0.4 |
GO:0032926 |
negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 |
0.4 |
GO:0010225 |
response to UV-C(GO:0010225) |
| 0.0 |
0.1 |
GO:1903061 |
regulation of protein lipidation(GO:1903059) positive regulation of protein lipidation(GO:1903061) |
| 0.0 |
0.3 |
GO:0051491 |
positive regulation of filopodium assembly(GO:0051491) |
| 0.0 |
0.3 |
GO:0090141 |
positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 |
1.8 |
GO:0070527 |
platelet aggregation(GO:0070527) |
| 0.0 |
0.3 |
GO:0006654 |
phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 |
0.2 |
GO:0042117 |
monocyte activation(GO:0042117) |
| 0.0 |
0.4 |
GO:1902018 |
negative regulation of cilium assembly(GO:1902018) |
| 0.0 |
1.1 |
GO:0046330 |
positive regulation of JNK cascade(GO:0046330) |
| 0.0 |
0.1 |
GO:0060729 |
intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 |
0.3 |
GO:0030206 |
chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 |
0.1 |
GO:2000501 |
natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.0 |
0.5 |
GO:0008209 |
androgen metabolic process(GO:0008209) |
| 0.0 |
0.3 |
GO:0048305 |
immunoglobulin secretion(GO:0048305) |
| 0.0 |
0.1 |
GO:1900119 |
positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 |
1.2 |
GO:0042073 |
intraciliary transport(GO:0042073) protein transport along microtubule(GO:0098840) |
| 0.0 |
0.6 |
GO:0016239 |
positive regulation of macroautophagy(GO:0016239) |
| 0.0 |
0.2 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.0 |
0.3 |
GO:0030539 |
male genitalia development(GO:0030539) |
| 0.0 |
0.8 |
GO:0006308 |
DNA catabolic process(GO:0006308) |
| 0.0 |
0.2 |
GO:0060398 |
regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 |
0.1 |
GO:1990253 |
cellular response to leucine starvation(GO:1990253) |
| 0.0 |
0.1 |
GO:0061462 |
protein localization to lysosome(GO:0061462) |
| 0.0 |
0.3 |
GO:1902187 |
negative regulation of viral release from host cell(GO:1902187) |
| 0.0 |
0.2 |
GO:0031666 |
detection of biotic stimulus(GO:0009595) positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) detection of molecule of bacterial origin(GO:0032490) detection of lipopolysaccharide(GO:0032497) detection of external biotic stimulus(GO:0098581) |
| 0.0 |
0.8 |
GO:1900046 |
regulation of blood coagulation(GO:0030193) regulation of hemostasis(GO:1900046) |
| 0.0 |
1.2 |
GO:0051453 |
regulation of intracellular pH(GO:0051453) |
| 0.0 |
0.3 |
GO:0030049 |
muscle filament sliding(GO:0030049) |
| 0.0 |
0.3 |
GO:0035871 |
protein K11-linked deubiquitination(GO:0035871) |
| 0.0 |
0.9 |
GO:0030968 |
endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 |
0.7 |
GO:0010829 |
negative regulation of glucose transport(GO:0010829) negative regulation of glucose import(GO:0046325) |
| 0.0 |
0.4 |
GO:0060384 |
innervation(GO:0060384) |
| 0.0 |
0.5 |
GO:0046580 |
negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 |
0.4 |
GO:0045879 |
negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 |
0.4 |
GO:0034198 |
cellular response to amino acid starvation(GO:0034198) |
| 0.0 |
0.2 |
GO:2000035 |
regulation of stem cell division(GO:2000035) |
| 0.0 |
0.4 |
GO:0055012 |
ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.0 |
2.8 |
GO:0035023 |
regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 |
0.8 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
0.2 |
GO:0035507 |
regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 |
0.2 |
GO:0032625 |
interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 |
0.6 |
GO:0000266 |
mitochondrial fission(GO:0000266) |
| 0.0 |
0.9 |
GO:0007029 |
endoplasmic reticulum organization(GO:0007029) |
| 0.0 |
0.5 |
GO:0006491 |
N-glycan processing(GO:0006491) |
| 0.0 |
1.7 |
GO:0000209 |
protein polyubiquitination(GO:0000209) |
| 0.0 |
4.5 |
GO:0043087 |
regulation of GTPase activity(GO:0043087) |
| 0.0 |
6.5 |
GO:0006897 |
endocytosis(GO:0006897) |
| 0.0 |
2.2 |
GO:0050714 |
positive regulation of protein secretion(GO:0050714) |
| 0.0 |
0.6 |
GO:0051180 |
vitamin transport(GO:0051180) |
| 0.0 |
0.2 |
GO:0070831 |
basement membrane assembly(GO:0070831) |
| 0.0 |
0.2 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.0 |
0.1 |
GO:0035964 |
COPI-coated vesicle budding(GO:0035964) |
| 0.0 |
0.2 |
GO:0009249 |
protein lipoylation(GO:0009249) |
| 0.0 |
0.5 |
GO:0007608 |
sensory perception of smell(GO:0007608) |
| 0.0 |
0.1 |
GO:0006172 |
ADP biosynthetic process(GO:0006172) |
| 0.0 |
0.2 |
GO:0031507 |
heterochromatin assembly(GO:0031507) |
| 0.0 |
0.4 |
GO:0045453 |
bone resorption(GO:0045453) |
| 0.0 |
0.0 |
GO:0043570 |
meiotic mismatch repair(GO:0000710) maintenance of DNA repeat elements(GO:0043570) |
| 0.0 |
0.3 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 |
0.1 |
GO:0036010 |
protein localization to endosome(GO:0036010) |
| 0.0 |
0.3 |
GO:0001570 |
vasculogenesis(GO:0001570) |
| 0.0 |
0.7 |
GO:0060395 |
SMAD protein signal transduction(GO:0060395) |
| 0.0 |
0.1 |
GO:0003094 |
glomerular filtration(GO:0003094) renal filtration(GO:0097205) |
| 0.0 |
0.1 |
GO:0017183 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 |
0.6 |
GO:0003333 |
amino acid transmembrane transport(GO:0003333) |
| 0.0 |
0.1 |
GO:0043388 |
positive regulation of DNA binding(GO:0043388) |
| 0.0 |
0.1 |
GO:0042554 |
superoxide anion generation(GO:0042554) |
| 0.0 |
5.3 |
GO:0007283 |
spermatogenesis(GO:0007283) |
| 0.0 |
0.3 |
GO:0006879 |
cellular iron ion homeostasis(GO:0006879) |
| 0.0 |
0.4 |
GO:0046854 |
phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 |
0.1 |
GO:0045947 |
negative regulation of translational initiation(GO:0045947) |
| 0.0 |
0.5 |
GO:0043473 |
pigmentation(GO:0043473) |
| 0.0 |
0.1 |
GO:0017144 |
drug metabolic process(GO:0017144) |
| 0.0 |
0.5 |
GO:0060997 |
dendritic spine morphogenesis(GO:0060997) |
| 0.0 |
0.5 |
GO:0046777 |
protein autophosphorylation(GO:0046777) |
| 0.0 |
0.3 |
GO:0001574 |
ganglioside biosynthetic process(GO:0001574) |
| 0.0 |
0.1 |
GO:0006741 |
NADP biosynthetic process(GO:0006741) |
| 0.0 |
0.0 |
GO:0032305 |
positive regulation of icosanoid secretion(GO:0032305) positive regulation of prostaglandin secretion(GO:0032308) |
| 0.0 |
0.2 |
GO:0043249 |
erythrocyte maturation(GO:0043249) |
| 0.0 |
0.2 |
GO:0051592 |
response to calcium ion(GO:0051592) |
| 0.0 |
0.4 |
GO:0008214 |
protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 |
0.2 |
GO:0006206 |
pyrimidine nucleobase metabolic process(GO:0006206) |
| 0.0 |
0.2 |
GO:0045653 |
negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 |
0.2 |
GO:0046579 |
positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 |
0.5 |
GO:0007584 |
response to nutrient(GO:0007584) |
| 0.0 |
0.7 |
GO:0099643 |
neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.0 |
0.1 |
GO:0019511 |
peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 |
0.2 |
GO:0042345 |
regulation of NF-kappaB import into nucleus(GO:0042345) NF-kappaB import into nucleus(GO:0042348) |
| 0.0 |
0.1 |
GO:0000055 |
ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 |
0.2 |
GO:0032981 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
0.1 |
GO:0036037 |
CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 |
0.3 |
GO:0008542 |
visual learning(GO:0008542) |
| 0.0 |
0.3 |
GO:0042475 |
odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 |
0.4 |
GO:0035914 |
skeletal muscle cell differentiation(GO:0035914) |
| 0.0 |
1.3 |
GO:0045444 |
fat cell differentiation(GO:0045444) |