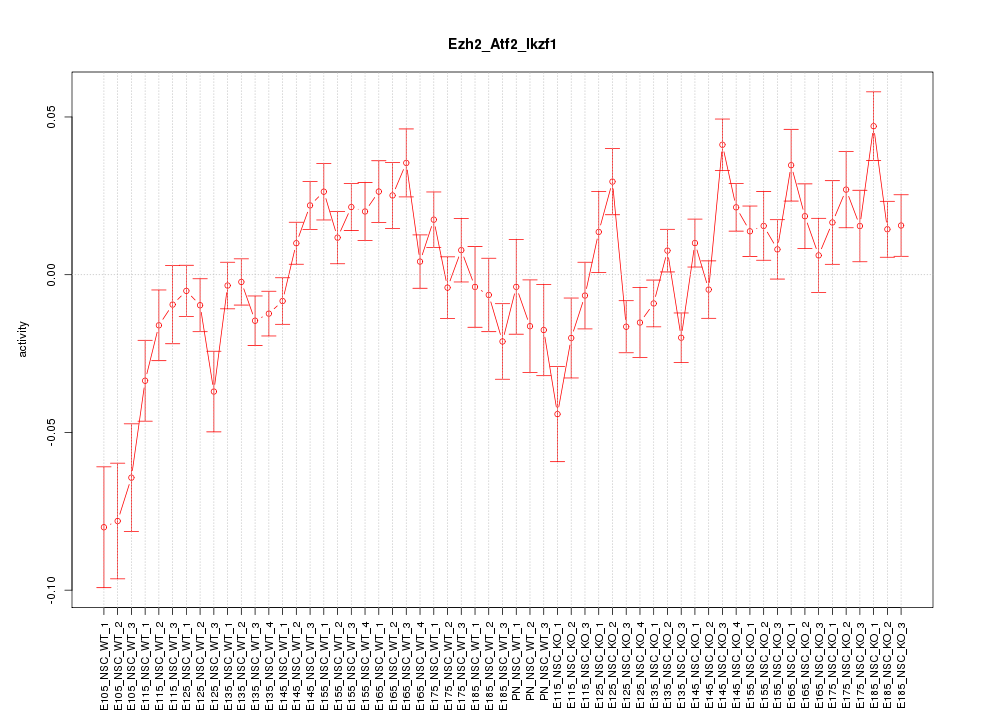
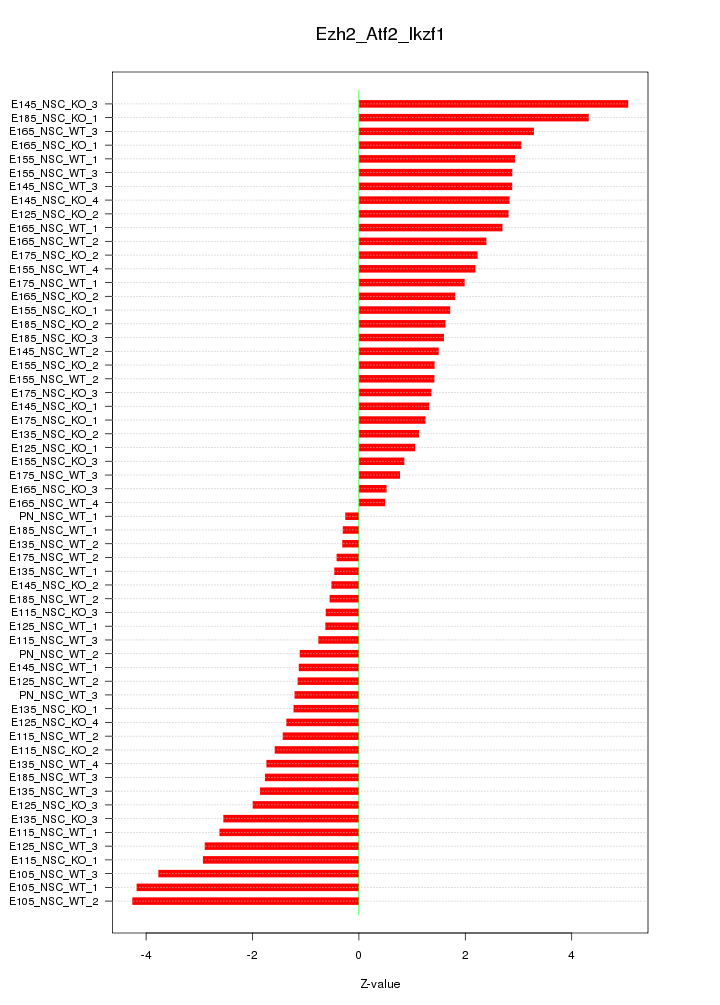
Motif ID: Ezh2_Atf2_Ikzf1
Z-value: 2.135

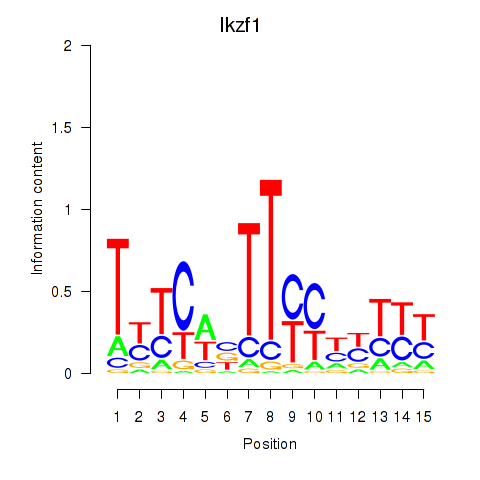
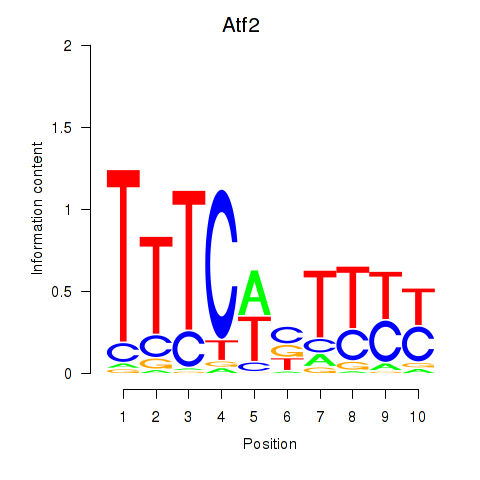
Transcription factors associated with Ezh2_Atf2_Ikzf1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Atf2 | ENSMUSG00000027104.12 | Atf2 |
| Ezh2 | ENSMUSG00000029687.10 | Ezh2 |
| Ikzf1 | ENSMUSG00000018654.11 | Ikzf1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Atf2 | mm10_v2_chr2_-_73892588_73892616 | 0.78 | 3.4e-13 | Click! |
| Ezh2 | mm10_v2_chr6_-_47594967_47595047 | -0.32 | 1.4e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.3 | 65.3 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 5.0 | 24.8 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 4.9 | 14.8 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 4.3 | 13.0 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 3.7 | 14.7 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 3.5 | 17.3 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 3.3 | 16.7 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 3.3 | 23.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 3.0 | 17.8 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 2.8 | 5.6 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 2.6 | 23.8 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 2.6 | 12.8 | GO:0021856 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 2.5 | 20.1 | GO:0097369 | sodium ion import(GO:0097369) |
| 2.5 | 12.5 | GO:0075136 | response to defenses of other organism involved in symbiotic interaction(GO:0052173) response to host defenses(GO:0052200) response to host(GO:0075136) |
| 2.4 | 9.5 | GO:0055099 | regulation of Cdc42 protein signal transduction(GO:0032489) response to high density lipoprotein particle(GO:0055099) platelet dense granule organization(GO:0060155) |
| 2.4 | 7.1 | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 2.3 | 11.5 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 2.3 | 6.8 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 2.1 | 29.4 | GO:0021540 | corpus callosum morphogenesis(GO:0021540) |
| 2.1 | 4.2 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 2.1 | 33.5 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 2.1 | 12.3 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 2.1 | 6.2 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 2.0 | 6.0 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 2.0 | 15.8 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 2.0 | 19.6 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 2.0 | 5.9 | GO:0003195 | tricuspid valve development(GO:0003175) tricuspid valve morphogenesis(GO:0003186) tricuspid valve formation(GO:0003195) |
| 1.9 | 7.7 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 1.9 | 11.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 1.9 | 7.4 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 1.8 | 7.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 1.8 | 7.1 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 1.8 | 5.3 | GO:0046880 | regulation of follicle-stimulating hormone secretion(GO:0046880) follicle-stimulating hormone secretion(GO:0046884) |
| 1.8 | 8.8 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 1.7 | 6.9 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 1.7 | 41.3 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 1.6 | 8.2 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 1.6 | 22.5 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 1.6 | 11.1 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 1.6 | 4.7 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 1.5 | 6.2 | GO:0097393 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 1.5 | 4.6 | GO:0044849 | estrous cycle(GO:0044849) |
| 1.5 | 10.6 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 1.5 | 13.6 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 1.5 | 3.0 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 1.5 | 55.4 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 1.4 | 4.3 | GO:1900736 | regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 1.4 | 5.7 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 1.4 | 9.8 | GO:0097090 | presynaptic membrane organization(GO:0097090) |
| 1.4 | 4.2 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 1.4 | 8.2 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 1.3 | 13.3 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 1.3 | 3.9 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 1.3 | 3.8 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 1.2 | 12.5 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 1.2 | 4.9 | GO:1903288 | regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 1.2 | 7.3 | GO:0019336 | phenol-containing compound catabolic process(GO:0019336) |
| 1.2 | 4.7 | GO:0021564 | glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 1.1 | 6.9 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 1.1 | 16.0 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 1.1 | 4.5 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 1.1 | 3.3 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 1.1 | 14.2 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 1.1 | 9.6 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 1.1 | 3.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 1.1 | 10.5 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 1.0 | 3.9 | GO:0032289 | central nervous system myelin formation(GO:0032289) cardiac cell fate specification(GO:0060912) |
| 1.0 | 3.9 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 1.0 | 3.8 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.9 | 13.1 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.9 | 33.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.9 | 10.2 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.9 | 0.9 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.9 | 1.8 | GO:2000790 | regulation of mesenchymal cell proliferation involved in lung development(GO:2000790) negative regulation of mesenchymal cell proliferation involved in lung development(GO:2000791) |
| 0.9 | 2.6 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.9 | 3.5 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.9 | 2.6 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.9 | 1.7 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.8 | 4.2 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.8 | 21.8 | GO:2000463 | positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.8 | 4.2 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.8 | 1.6 | GO:2000173 | negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.8 | 4.8 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.8 | 7.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.8 | 8.6 | GO:1903421 | regulation of synaptic vesicle recycling(GO:1903421) |
| 0.8 | 6.2 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.8 | 32.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.8 | 2.3 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.8 | 6.2 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.8 | 55.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.8 | 1.5 | GO:0010958 | regulation of amino acid import(GO:0010958) regulation of L-arginine import(GO:0010963) |
| 0.7 | 4.4 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.7 | 7.3 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.7 | 2.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.7 | 15.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.7 | 1.4 | GO:0002730 | regulation of dendritic cell cytokine production(GO:0002730) |
| 0.7 | 1.3 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.7 | 2.0 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.7 | 7.9 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.6 | 4.5 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.6 | 52.6 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.6 | 4.5 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.6 | 1.9 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.6 | 1.9 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.6 | 17.6 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.6 | 2.4 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.6 | 1.8 | GO:1901731 | calcium-mediated signaling using extracellular calcium source(GO:0035585) positive regulation of platelet aggregation(GO:1901731) |
| 0.6 | 8.2 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.6 | 4.1 | GO:0034047 | regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.6 | 8.0 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.6 | 1.7 | GO:0048143 | astrocyte activation(GO:0048143) |
| 0.6 | 3.9 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.5 | 1.6 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.5 | 2.2 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.5 | 2.7 | GO:0032911 | nerve growth factor production(GO:0032902) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.5 | 1.6 | GO:0046958 | nonassociative learning(GO:0046958) |
| 0.5 | 4.7 | GO:0047496 | vesicle transport along microtubule(GO:0047496) |
| 0.5 | 2.1 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.5 | 5.1 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.5 | 4.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.5 | 1.0 | GO:0014054 | regulation of gamma-aminobutyric acid secretion(GO:0014052) positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.5 | 2.0 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.5 | 2.5 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.5 | 2.0 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.5 | 2.0 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.5 | 2.9 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.5 | 1.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.5 | 1.4 | GO:0031133 | regulation of axon diameter(GO:0031133) intermediate filament bundle assembly(GO:0045110) |
| 0.5 | 19.5 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.5 | 0.9 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.5 | 7.5 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.5 | 1.4 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.5 | 0.5 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.5 | 33.8 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.5 | 1.4 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.4 | 1.7 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.4 | 3.8 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.4 | 2.9 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.4 | 15.0 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.4 | 19.5 | GO:0008542 | visual learning(GO:0008542) |
| 0.4 | 1.6 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.4 | 1.6 | GO:0071442 | regulation of histone H3-K14 acetylation(GO:0071440) positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.4 | 16.0 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.4 | 1.6 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.4 | 1.5 | GO:0031652 | positive regulation of heat generation(GO:0031652) |
| 0.4 | 1.5 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) |
| 0.4 | 1.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.4 | 1.9 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.4 | 2.6 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.4 | 0.7 | GO:1902947 | regulation of tau-protein kinase activity(GO:1902947) |
| 0.4 | 1.4 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.4 | 1.4 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.4 | 1.8 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.4 | 2.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.4 | 25.4 | GO:0007612 | learning(GO:0007612) |
| 0.3 | 1.0 | GO:0071476 | cellular hypotonic response(GO:0071476) |
| 0.3 | 2.1 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.3 | 1.0 | GO:0060745 | mammary gland branching involved in pregnancy(GO:0060745) |
| 0.3 | 2.4 | GO:0001964 | startle response(GO:0001964) |
| 0.3 | 1.7 | GO:1901678 | iron coordination entity transport(GO:1901678) |
| 0.3 | 7.5 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.3 | 5.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.3 | 5.9 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.3 | 2.0 | GO:0002524 | hypersensitivity(GO:0002524) |
| 0.3 | 2.0 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.3 | 0.7 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.3 | 5.5 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.3 | 1.0 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.3 | 4.8 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.3 | 2.8 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.3 | 1.6 | GO:0002674 | negative regulation of acute inflammatory response(GO:0002674) |
| 0.3 | 1.9 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.3 | 0.9 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.3 | 0.9 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.3 | 1.5 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.3 | 1.5 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.3 | 0.9 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.3 | 0.9 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.3 | 1.8 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.3 | 1.2 | GO:0044330 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.3 | 9.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.3 | 2.3 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.3 | 8.3 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.3 | 0.9 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.3 | 2.8 | GO:0006691 | leukotriene metabolic process(GO:0006691) |
| 0.3 | 3.1 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.3 | 1.1 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.3 | 0.8 | GO:1902950 | regulation of dendritic spine maintenance(GO:1902950) positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.3 | 6.8 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.3 | 1.6 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.3 | 1.3 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.3 | 1.0 | GO:0034727 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) late nucleophagy(GO:0044805) single-organism membrane invagination(GO:1902534) |
| 0.3 | 9.4 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.3 | 1.8 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.3 | 3.3 | GO:0097484 | dendrite extension(GO:0097484) |
| 0.3 | 0.8 | GO:1902219 | regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.3 | 0.8 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.2 | 0.5 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.2 | 2.7 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.2 | 12.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.2 | 4.4 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
| 0.2 | 4.4 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.2 | 1.5 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.2 | 1.2 | GO:0014842 | regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.2 | 3.7 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.2 | 8.2 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.2 | 0.5 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.2 | 1.4 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 0.7 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.2 | 1.4 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.2 | 3.4 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.2 | 2.0 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.2 | 0.9 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.2 | 2.7 | GO:1900273 | positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.2 | 1.5 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.2 | 1.5 | GO:0050807 | regulation of synapse organization(GO:0050807) |
| 0.2 | 1.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.2 | 2.3 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.2 | 2.5 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.2 | 0.4 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) |
| 0.2 | 2.3 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.2 | 2.7 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.2 | 2.0 | GO:0001504 | neurotransmitter uptake(GO:0001504) |
| 0.2 | 1.0 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.2 | 1.4 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.2 | 2.2 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.2 | 1.8 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.2 | 4.7 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.2 | 20.9 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.2 | 0.8 | GO:0071105 | macrophage activation involved in immune response(GO:0002281) response to interleukin-11(GO:0071105) osteoclast fusion(GO:0072675) |
| 0.2 | 3.3 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 2.1 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.2 | 2.3 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.2 | 1.5 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.2 | 1.1 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.2 | 9.8 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.2 | 5.8 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.2 | 0.6 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.2 | 12.0 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.2 | 8.2 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.2 | 4.0 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.2 | 0.7 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.2 | 0.9 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.2 | 16.6 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.2 | 1.1 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.2 | 2.5 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.2 | 0.4 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.2 | 0.7 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.2 | 2.6 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.2 | 1.4 | GO:0036506 | maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.2 | 0.7 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.2 | 0.7 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.2 | 3.0 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.2 | 2.1 | GO:0014898 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.2 | 2.3 | GO:0009072 | aromatic amino acid family metabolic process(GO:0009072) |
| 0.2 | 1.0 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.2 | 1.1 | GO:0071941 | nitrate metabolic process(GO:0042126) nitrogen cycle metabolic process(GO:0071941) |
| 0.2 | 1.6 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.2 | 2.2 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.2 | 0.6 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.2 | 0.6 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.2 | 0.5 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.2 | 0.2 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.2 | 1.2 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 | 0.9 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 4.8 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 1.3 | GO:0002192 | cap-independent translational initiation(GO:0002190) IRES-dependent translational initiation(GO:0002192) |
| 0.1 | 1.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 4.7 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 7.6 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 0.6 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 0.4 | GO:0090285 | regulation of protein glycosylation in Golgi(GO:0090283) negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.1 | 0.6 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.8 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.1 | 1.2 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.1 | 0.7 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.1 | 0.7 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 1.2 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 1.9 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.1 | 1.3 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 1.8 | GO:0001553 | luteinization(GO:0001553) |
| 0.1 | 1.4 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.1 | 0.5 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 0.3 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.1 | 1.3 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.9 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 0.5 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 1.8 | GO:0044091 | membrane biogenesis(GO:0044091) |
| 0.1 | 2.5 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 2.0 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 1.8 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 1.3 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 | 0.3 | GO:0010624 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.1 | 0.7 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 1.3 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 0.8 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.4 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 1.4 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.6 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 0.6 | GO:0031442 | positive regulation of mRNA 3'-end processing(GO:0031442) |
| 0.1 | 2.4 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.7 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.5 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 | 1.8 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 | 0.2 | GO:0010847 | regulation of chromatin assembly(GO:0010847) negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.1 | 1.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.7 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 0.4 | GO:0072429 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 | 0.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 0.9 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.1 | 0.8 | GO:0030813 | positive regulation of nucleotide catabolic process(GO:0030813) positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.1 | 2.8 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.1 | 0.8 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.2 | GO:2000848 | positive regulation of corticosteroid hormone secretion(GO:2000848) |
| 0.1 | 0.3 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 2.2 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.1 | 0.5 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 0.7 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.1 | 0.9 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 1.9 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.1 | 0.5 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 0.4 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.1 | 0.7 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.1 | 0.8 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.1 | 0.4 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.6 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 1.4 | GO:0048741 | skeletal muscle fiber development(GO:0048741) |
| 0.1 | 1.5 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 0.9 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 0.8 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 0.1 | GO:0010881 | regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion(GO:0010881) |
| 0.1 | 0.9 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.1 | 0.7 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 4.5 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.1 | 0.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.5 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.1 | 7.9 | GO:0006333 | chromatin assembly or disassembly(GO:0006333) |
| 0.1 | 0.9 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.1 | 1.4 | GO:0015991 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.1 | 0.3 | GO:0048284 | organelle fusion(GO:0048284) |
| 0.1 | 0.3 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 0.3 | GO:0034116 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.1 | 1.6 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.1 | 0.3 | GO:0045162 | neuronal ion channel clustering(GO:0045161) clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 2.2 | GO:0048489 | synaptic vesicle transport(GO:0048489) establishment of synaptic vesicle localization(GO:0097480) |
| 0.0 | 0.4 | GO:0051882 | mitochondrial depolarization(GO:0051882) |
| 0.0 | 0.6 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.0 | 0.4 | GO:0005980 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.9 | GO:0034340 | response to type I interferon(GO:0034340) |
| 0.0 | 1.1 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 0.2 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.6 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.4 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.4 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.6 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 1.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 1.0 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.6 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.4 | GO:0043242 | negative regulation of actin filament depolymerization(GO:0030835) negative regulation of protein complex disassembly(GO:0043242) |
| 0.0 | 0.7 | GO:0051101 | regulation of DNA binding(GO:0051101) |
| 0.0 | 0.3 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.4 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 | 4.7 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 1.6 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 0.1 | GO:2000727 | positive regulation of cardiac muscle cell differentiation(GO:2000727) |
| 0.0 | 1.5 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.3 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.0 | 1.2 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 0.4 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 0.6 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.3 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 3.5 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.4 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 1.0 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 0.8 | GO:0006497 | protein lipidation(GO:0006497) |
| 0.0 | 0.2 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0060546 | negative regulation of necroptotic process(GO:0060546) |
| 0.0 | 1.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.1 | GO:0033004 | negative regulation of mast cell activation(GO:0033004) |
| 0.0 | 0.1 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.9 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 0.8 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.2 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.3 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.7 | GO:0007611 | learning or memory(GO:0007611) |
| 0.0 | 0.1 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.2 | GO:0009268 | response to pH(GO:0009268) |
| 0.0 | 0.0 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.0 | GO:0008228 | response to yeast(GO:0001878) opsonization(GO:0008228) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 46.6 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 4.7 | 18.8 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 3.4 | 70.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 2.9 | 11.5 | GO:0043511 | inhibin complex(GO:0043511) |
| 2.4 | 7.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 2.3 | 14.0 | GO:0044308 | axonal spine(GO:0044308) |
| 2.2 | 11.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 2.0 | 34.7 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 2.0 | 39.2 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 1.8 | 16.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 1.5 | 6.2 | GO:1990421 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 1.5 | 19.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 1.5 | 4.6 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 1.5 | 10.5 | GO:0071437 | invadopodium(GO:0071437) |
| 1.5 | 23.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 1.3 | 88.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 1.2 | 2.3 | GO:0044307 | dendritic branch(GO:0044307) |
| 1.1 | 8.0 | GO:0005883 | neurofilament(GO:0005883) |
| 1.1 | 12.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 1.0 | 5.2 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 1.0 | 9.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 1.0 | 42.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 1.0 | 3.9 | GO:0071797 | LUBAC complex(GO:0071797) |
| 1.0 | 2.9 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.9 | 14.4 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.9 | 4.3 | GO:0033648 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.8 | 15.2 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.8 | 3.3 | GO:0031673 | H zone(GO:0031673) |
| 0.8 | 0.8 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.8 | 6.9 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.7 | 4.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.7 | 2.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.7 | 4.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.7 | 18.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.7 | 2.0 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.6 | 12.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.6 | 1.8 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.6 | 10.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.5 | 3.8 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.5 | 2.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.5 | 4.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.5 | 7.2 | GO:0043203 | axon hillock(GO:0043203) |
| 0.5 | 12.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.5 | 0.5 | GO:0044299 | C-fiber(GO:0044299) |
| 0.5 | 30.0 | GO:0030315 | T-tubule(GO:0030315) |
| 0.5 | 8.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.4 | 1.8 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.4 | 1.8 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.4 | 1.7 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.4 | 3.2 | GO:0030175 | filopodium(GO:0030175) |
| 0.4 | 1.2 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.4 | 11.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.3 | 36.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.3 | 3.3 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.3 | 10.9 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.3 | 3.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.3 | 2.9 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.3 | 3.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.3 | 1.5 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.3 | 1.5 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.3 | 2.7 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.3 | 9.1 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.3 | 1.4 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.3 | 1.1 | GO:0043259 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) |
| 0.3 | 1.7 | GO:0005775 | vacuolar lumen(GO:0005775) |
| 0.3 | 3.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.3 | 6.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.3 | 2.9 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.3 | 52.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.3 | 4.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.3 | 1.8 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.3 | 6.1 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.2 | 21.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.2 | 10.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 19.1 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.2 | 5.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.2 | 4.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 13.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.2 | 1.5 | GO:0033503 | HULC complex(GO:0033503) |
| 0.2 | 5.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.2 | 0.6 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.2 | 1.4 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.2 | 3.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 16.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 2.4 | GO:0042581 | specific granule(GO:0042581) |
| 0.2 | 1.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.2 | 2.0 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 1.6 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.2 | 24.2 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.2 | 5.2 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 3.0 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 0.3 | GO:0012506 | vesicle membrane(GO:0012506) |
| 0.1 | 4.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 0.4 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 3.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.9 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 2.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 1.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 5.6 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 7.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 3.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 0.7 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 2.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 3.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 3.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 13.4 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 0.3 | GO:0031230 | intrinsic component of cell outer membrane(GO:0031230) integral component of cell outer membrane(GO:0045203) |
| 0.1 | 21.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 1.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.7 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.1 | 0.5 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 4.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 2.9 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 0.9 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.1 | 0.8 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 0.4 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 6.6 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.1 | 0.7 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 1.1 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 0.7 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.1 | 3.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 0.5 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 2.3 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 0.5 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 2.1 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 0.7 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 1.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 0.4 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 3.0 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.1 | 0.7 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 0.4 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 1.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 2.7 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 3.9 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.1 | 2.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 4.8 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 0.3 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.1 | 0.3 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 0.3 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 0.3 | GO:0070160 | occluding junction(GO:0070160) |
| 0.0 | 1.3 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 1.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.3 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.1 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.0 | 3.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.8 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 2.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.6 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.3 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.5 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 1.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 84.7 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.3 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.4 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 4.6 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 45.5 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 5.1 | 20.4 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 4.8 | 33.9 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 4.1 | 12.2 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 3.3 | 13.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 3.2 | 15.9 | GO:0033691 | sialic acid binding(GO:0033691) |
| 3.2 | 9.5 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) |
| 3.0 | 18.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 3.0 | 18.0 | GO:0019841 | retinol binding(GO:0019841) |
| 2.5 | 29.9 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 2.5 | 7.4 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 2.4 | 9.4 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 2.3 | 18.3 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 2.2 | 13.4 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 2.1 | 8.5 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 2.0 | 34.7 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 2.0 | 27.9 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 1.8 | 5.4 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 1.8 | 16.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 1.8 | 8.8 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 1.7 | 6.9 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 1.7 | 13.6 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 1.6 | 8.2 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 1.6 | 6.4 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 1.5 | 6.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 1.4 | 34.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 1.4 | 4.3 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 1.4 | 49.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 1.4 | 7.0 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 1.4 | 4.2 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 1.3 | 10.7 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 1.3 | 6.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 1.3 | 3.9 | GO:0031752 | D3 dopamine receptor binding(GO:0031750) D5 dopamine receptor binding(GO:0031752) |
| 1.3 | 5.1 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 1.2 | 7.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 1.2 | 5.8 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 1.1 | 10.0 | GO:0034711 | inhibin binding(GO:0034711) |
| 1.1 | 3.2 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 1.1 | 3.2 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 1.0 | 6.8 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 1.0 | 2.9 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.9 | 7.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.9 | 6.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.9 | 6.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.9 | 4.4 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.9 | 2.6 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.9 | 2.6 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.9 | 11.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.9 | 10.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.8 | 6.8 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.8 | 15.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.8 | 17.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.8 | 5.5 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.8 | 2.3 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.8 | 2.3 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.7 | 2.2 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.7 | 14.4 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.7 | 5.0 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.7 | 2.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.7 | 2.9 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.7 | 2.8 | GO:0001639 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) |
| 0.7 | 19.0 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.7 | 11.9 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.7 | 2.1 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.7 | 3.5 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.7 | 2.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.7 | 5.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.6 | 5.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.6 | 4.3 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.6 | 1.8 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.6 | 5.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.6 | 13.0 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.6 | 2.8 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.6 | 1.7 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.5 | 3.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.5 | 4.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.5 | 2.7 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.5 | 2.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.5 | 4.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.5 | 4.5 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.5 | 36.5 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.5 | 2.0 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.5 | 2.0 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.5 | 1.9 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.5 | 4.7 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.5 | 2.8 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.5 | 5.6 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.5 | 11.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.5 | 1.4 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.5 | 3.2 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.5 | 3.6 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.5 | 1.4 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.5 | 1.4 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.4 | 1.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.4 | 6.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.4 | 6.1 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.4 | 1.7 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.4 | 3.2 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.4 | 0.8 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.4 | 2.0 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.4 | 1.1 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.4 | 2.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.4 | 3.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.4 | 7.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.4 | 3.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.3 | 2.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.3 | 1.0 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.3 | 1.4 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.3 | 2.4 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.3 | 2.7 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.3 | 23.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.3 | 2.0 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.3 | 2.9 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.3 | 15.4 | GO:0004120 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.3 | 2.5 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.3 | 2.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.3 | 4.1 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.3 | 1.7 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.3 | 1.4 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.3 | 1.7 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.3 | 5.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.3 | 0.8 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.3 | 2.8 | GO:0042301 | phosphate ion binding(GO:0042301) inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.3 | 4.2 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.3 | 2.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.3 | 0.8 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.3 | 1.5 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.2 | 1.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.2 | 1.5 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 1.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.2 | 3.1 | GO:0008748 | N-ethylmaleimide reductase activity(GO:0008748) reduced coenzyme F420 dehydrogenase activity(GO:0043738) sulfur oxygenase reductase activity(GO:0043826) malolactic enzyme activity(GO:0043883) NADPH:sulfur oxidoreductase activity(GO:0043914) epoxyqueuosine reductase activity(GO:0052693) |
| 0.2 | 1.9 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.2 | 4.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.2 | 0.9 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.2 | 0.9 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.2 | 1.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) lytic endotransglycosylase activity(GO:0008932) |
| 0.2 | 1.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.2 | 1.5 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.2 | 0.8 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.2 | 3.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.2 | 4.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 0.8 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.2 | 0.8 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.2 | 3.6 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.2 | 0.6 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.2 | 3.7 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.2 | 3.9 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.2 | 22.2 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.2 | 0.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.2 | 1.0 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.2 | 1.6 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 0.7 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 4.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 0.8 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 1.8 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.2 | 4.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 1.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.2 | 5.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 2.9 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.2 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 0.2 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.2 | 1.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.2 | 2.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.2 | 18.2 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.2 | 0.8 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.2 | 0.5 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.2 | 1.8 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.9 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.1 | 1.6 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 1.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.9 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.1 | 2.6 | GO:0044390 | ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.1 | 0.9 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 45.3 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 1.5 | GO:0034943 | acyl-CoA ligase activity(GO:0003996) 3-oxo-2-(2'-pentenyl)cyclopentane-1-octanoic acid CoA ligase activity(GO:0010435) 3-isopropenyl-6-oxoheptanoyl-CoA synthetase activity(GO:0018854) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA synthetase activity(GO:0018855) benzoyl acetate-CoA ligase activity(GO:0018856) 2,4-dichlorobenzoate-CoA ligase activity(GO:0018857) pivalate-CoA ligase activity(GO:0034783) cyclopropanecarboxylate-CoA ligase activity(GO:0034793) adipate-CoA ligase activity(GO:0034796) citronellyl-CoA ligase activity(GO:0034823) mentha-1,3-dione-CoA ligase activity(GO:0034841) thiophene-2-carboxylate-CoA ligase activity(GO:0034842) 2,4,4-trimethylpentanoate-CoA ligase activity(GO:0034865) cis-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034942) trans-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034943) branched-chain acyl-CoA synthetase (ADP-forming) activity(GO:0043759) aryl-CoA synthetase (ADP-forming) activity(GO:0043762) 3-hydroxypropionyl-CoA synthetase activity(GO:0043955) perillic acid:CoA ligase (ADP-forming) activity(GO:0052685) perillic acid:CoA ligase (AMP-forming) activity(GO:0052686) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (ADP-forming) activity(GO:0052687) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (AMP-forming) activity(GO:0052688) pristanate-CoA ligase activity(GO:0070251) malonyl-CoA synthetase activity(GO:0090409) |
| 0.1 | 1.0 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 0.7 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 1.9 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 4.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.8 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 2.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 5.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 0.7 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 7.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 1.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 1.9 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 3.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 2.9 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.4 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.6 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 2.7 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 0.5 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.1 | 0.6 | GO:0032407 | MutSalpha complex binding(GO:0032407) |
| 0.1 | 0.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 3.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 1.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 1.8 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.1 | 0.3 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 12.6 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 0.4 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 1.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.9 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 1.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 4.2 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 2.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 11.0 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 0.2 | GO:0004942 | anaphylatoxin receptor activity(GO:0004942) |
| 0.1 | 0.8 | GO:0017161 | protein tyrosine/threonine phosphatase activity(GO:0008330) JUN kinase phosphatase activity(GO:0008579) phosphohistidine phosphatase activity(GO:0008969) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) NADP phosphatase activity(GO:0019178) transmembrane receptor protein phosphatase activity(GO:0019198) 5-amino-6-(5-phosphoribitylamino)uracil phosphatase activity(GO:0043726) phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) phosphatidylinositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052867) IDP phosphatase activity(GO:1990003) |
| 0.1 | 4.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 0.9 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 1.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 0.4 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.4 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 1.4 | GO:0051787 | misfolded protein binding(GO:0051787) proteasome binding(GO:0070628) ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 5.0 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.1 | 0.4 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 9.4 | GO:0003729 | mRNA binding(GO:0003729) |
| 0.1 | 1.1 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.1 | 1.1 | GO:0015085 | calcium channel activity(GO:0005262) calcium ion transmembrane transporter activity(GO:0015085) |
| 0.1 | 0.6 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 1.3 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 0.9 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.3 | GO:0043842 | Kdo transferase activity(GO:0043842) |
| 0.1 | 23.8 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 0.2 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.1 | 0.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 17.3 | GO:0043774 | UDP-N-acetylmuramoylalanyl-D-glutamyl-2,6-diaminopimelate-D-alanyl-D-alanine ligase activity(GO:0008766) ribosomal S6-glutamic acid ligase activity(GO:0018169) coenzyme F420-0 gamma-glutamyl ligase activity(GO:0043773) coenzyme F420-2 alpha-glutamyl ligase activity(GO:0043774) protein-glycine ligase activity(GO:0070735) protein-glycine ligase activity, initiating(GO:0070736) protein-glycine ligase activity, elongating(GO:0070737) tubulin-glycine ligase activity(GO:0070738) protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 1.2 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 1.0 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 1.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 1.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 1.4 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 0.9 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.1 | 2.8 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.1 | 0.7 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) |
| 0.0 | 0.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.6 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 1.2 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 1.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 1.4 | GO:0018602 | sulfonate dioxygenase activity(GO:0000907) 2,4-dichlorophenoxyacetate alpha-ketoglutarate dioxygenase activity(GO:0018602) hypophosphite dioxygenase activity(GO:0034792) gibberellin 2-beta-dioxygenase activity(GO:0045543) C-19 gibberellin 2-beta-dioxygenase activity(GO:0052634) C-20 gibberellin 2-beta-dioxygenase activity(GO:0052635) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0045703 | pinocarveol dehydrogenase activity(GO:0018446) chloral hydrate dehydrogenase activity(GO:0018447) hydroxymethylmethylsilanediol oxidase activity(GO:0018448) 1-phenylethanol dehydrogenase activity(GO:0018449) myrtenol dehydrogenase activity(GO:0018450) cis-1,2-dihydroxy-1,2-dihydro-8-carboxynaphthalene dehydrogenase activity(GO:0034522) 3-hydroxy-4-methyloctanoyl-CoA dehydrogenase activity(GO:0034582) 2-hydroxy-4-isopropenylcyclohexane-1-carboxyl-CoA dehydrogenase activity(GO:0034778) cis-9,10-dihydroanthracene-9,10-diol dehydrogenase activity(GO:0034817) citronellol dehydrogenase activity(GO:0034821) naphthyl-2-hydroxymethyl-succinyl-CoA dehydrogenase activity(GO:0034847) 2,4,4-trimethyl-1-pentanol dehydrogenase activity(GO:0034863) 2,4,4-trimethyl-3-hydroxypentanoyl-CoA dehydrogenase activity(GO:0034868) 1-hydroxy-4,4-dimethylpentan-3-one dehydrogenase activity(GO:0034871) endosulfan diol dehydrogenase activity(GO:0034891) endosulfan hydroxyether dehydrogenase activity(GO:0034901) 3-hydroxy-2-methylhexanoyl-CoA dehydrogenase activity(GO:0034918) 3-hydroxy-2,6-dimethyl-5-methylene-heptanoyl-CoA dehydrogenase activity(GO:0034944) versicolorin reductase activity(GO:0042469) ketoreductase activity(GO:0045703) |
| 0.0 | 2.1 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.0 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 0.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 1.6 | GO:0008514 | organic anion transmembrane transporter activity(GO:0008514) |
| 0.0 | 0.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 2.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.2 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.0 | 1.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.4 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 1.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 2.7 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.5 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.0 | 1.5 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.0 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.0 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.0 | 0.3 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.1 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.0 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |