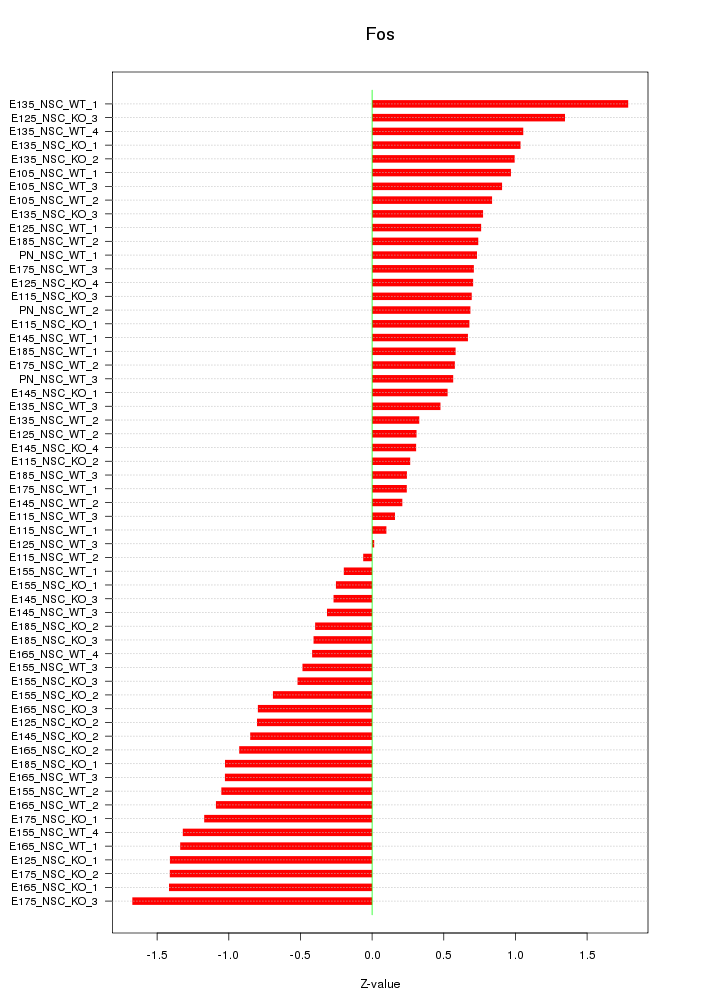
Motif ID: Fos
Z-value: 0.829
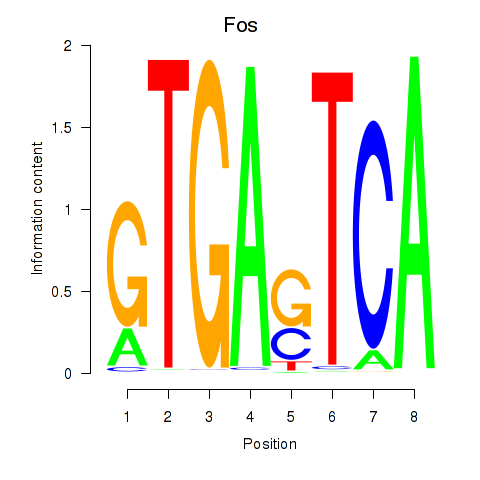
Transcription factors associated with Fos:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Fos | ENSMUSG00000021250.7 | Fos |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Fos | mm10_v2_chr12_+_85473883_85473896 | 0.05 | 7.1e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.2 | GO:0060364 | frontal suture morphogenesis(GO:0060364) |
| 1.9 | 5.7 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 1.7 | 5.2 | GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 1.4 | 13.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 1.3 | 4.0 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 1.3 | 1.3 | GO:0034113 | heterotypic cell-cell adhesion(GO:0034113) |
| 1.2 | 4.9 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 1.1 | 4.3 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 1.0 | 5.1 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.9 | 3.7 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.9 | 4.5 | GO:0021764 | amygdala development(GO:0021764) |
| 0.9 | 2.7 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.8 | 2.5 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 0.8 | 2.4 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.7 | 2.1 | GO:0030300 | regulation of intestinal cholesterol absorption(GO:0030300) |
| 0.7 | 3.4 | GO:0015705 | iodide transport(GO:0015705) |
| 0.7 | 1.3 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.5 | 1.6 | GO:0097278 | complement-dependent cytotoxicity(GO:0097278) |
| 0.5 | 1.5 | GO:2000851 | positive regulation of glucocorticoid secretion(GO:2000851) regulation of corticosterone secretion(GO:2000852) |
| 0.5 | 1.0 | GO:0002295 | T-helper cell lineage commitment(GO:0002295) |
| 0.5 | 2.4 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.5 | 4.2 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.4 | 1.8 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.4 | 2.0 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.4 | 3.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.4 | 1.5 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.4 | 2.3 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.4 | 1.8 | GO:0046133 | pyrimidine ribonucleoside catabolic process(GO:0046133) |
| 0.4 | 8.9 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.3 | 2.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.3 | 3.5 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.3 | 2.9 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
| 0.3 | 0.9 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.3 | 1.8 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.3 | 8.3 | GO:0009409 | response to cold(GO:0009409) |
| 0.3 | 0.9 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.3 | 3.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.3 | 2.8 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.3 | 0.8 | GO:0007621 | negative regulation of female receptivity(GO:0007621) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of cellular pH reduction(GO:0032847) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.3 | 5.0 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.3 | 3.9 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.3 | 0.5 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.3 | 1.0 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.3 | 5.5 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.2 | 2.5 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.2 | 4.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.2 | 0.7 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.2 | 2.7 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.2 | 0.7 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) |
| 0.2 | 1.3 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 0.2 | 1.3 | GO:0036233 | glycine import(GO:0036233) |
| 0.2 | 4.8 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.2 | 4.6 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.2 | 1.6 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.2 | 1.8 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.2 | 3.4 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.2 | 3.3 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.2 | 2.0 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.2 | 2.9 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.2 | 4.6 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.2 | 1.9 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.2 | 3.1 | GO:0048535 | lymph node development(GO:0048535) |
| 0.2 | 0.8 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.2 | 2.6 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.2 | 4.2 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.2 | 0.5 | GO:0048388 | endosomal lumen acidification(GO:0048388) synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 | 2.8 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.1 | 1.0 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 0.7 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.1 | 4.1 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.1 | 1.2 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.1 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.1 | 1.9 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.1 | 1.5 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 1.7 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.1 | 1.4 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.8 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.1 | 1.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 0.8 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.4 | GO:1904451 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.1 | 0.5 | GO:0090370 | negative regulation of sterol transport(GO:0032372) negative regulation of cholesterol transport(GO:0032375) negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 | 1.8 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.1 | 5.2 | GO:0001938 | positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.1 | 0.4 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 1.8 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 2.4 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.1 | 1.7 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.1 | 0.3 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 1.3 | GO:0061000 | negative regulation of dendritic spine development(GO:0061000) |
| 0.1 | 2.8 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 1.6 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 0.5 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 1.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 2.8 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 1.2 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.1 | 0.8 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 0.5 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 | 0.6 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.1 | 5.5 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 0.9 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 2.9 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.1 | 1.1 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.1 | 3.3 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 3.6 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.1 | 0.5 | GO:0071557 | notochord morphogenesis(GO:0048570) histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 2.0 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.1 | 0.7 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.1 | 0.2 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.1 | 0.9 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 0.6 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 0.5 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.1 | 3.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 4.1 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.3 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 1.0 | GO:0048678 | response to axon injury(GO:0048678) |
| 0.0 | 0.9 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.1 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 | 1.6 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 2.2 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.0 | 0.4 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 4.6 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.6 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 0.7 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.0 | 1.4 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.2 | GO:0061470 | interleukin-21 production(GO:0032625) T follicular helper cell differentiation(GO:0061470) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.2 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.5 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.0 | 1.0 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 3.0 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 2.4 | GO:0030833 | regulation of actin filament polymerization(GO:0030833) |
| 0.0 | 0.2 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.2 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 0.9 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.5 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 1.4 | GO:1903749 | positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 1.0 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 4.4 | GO:0043086 | negative regulation of catalytic activity(GO:0043086) |
| 0.0 | 0.3 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 0.4 | GO:0006220 | pyrimidine nucleotide metabolic process(GO:0006220) |
| 0.0 | 4.4 | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process(GO:0043161) |
| 0.0 | 0.0 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.0 | 1.4 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:1990047 | spindle matrix(GO:1990047) |
| 1.3 | 5.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 1.0 | 13.8 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.6 | 1.8 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.6 | 2.4 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.6 | 1.8 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 0.5 | 1.4 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.4 | 4.9 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.4 | 1.3 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.4 | 3.0 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.4 | 2.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.3 | 2.4 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.3 | 1.7 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.3 | 1.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.3 | 4.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.3 | 2.7 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.3 | 3.3 | GO:0031105 | septin complex(GO:0031105) |
| 0.3 | 2.5 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.3 | 2.6 | GO:0098533 | ATPase dependent transmembrane transport complex(GO:0098533) |
| 0.2 | 12.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 4.0 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.2 | 1.6 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.2 | 2.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 1.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 3.2 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.2 | 1.0 | GO:0008623 | CHRAC(GO:0008623) |
| 0.2 | 0.8 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.2 | 6.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 3.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 6.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 3.1 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 2.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 3.5 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 2.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 1.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 5.5 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 0.4 | GO:1990795 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) |
| 0.1 | 0.3 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 0.7 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 1.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 4.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 1.1 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 1.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.5 | GO:0044613 | nuclear pore central transport channel(GO:0044613) Flemming body(GO:0090543) |
| 0.1 | 2.0 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 6.8 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.1 | 0.5 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 3.5 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 0.5 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.8 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.0 | 2.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 2.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 2.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.4 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 2.8 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 1.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 3.3 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 0.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 7.1 | GO:0005924 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.0 | 2.3 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 2.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.7 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 13.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 1.2 | 4.9 | GO:1990254 | keratin filament binding(GO:1990254) |
| 1.0 | 4.8 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 1.0 | 3.8 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.9 | 3.4 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.8 | 3.4 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.8 | 5.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.8 | 4.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.8 | 4.6 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.5 | 2.5 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.5 | 2.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.5 | 2.9 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.5 | 1.8 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) phospholipase A2 inhibitor activity(GO:0019834) |
| 0.4 | 1.8 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.4 | 8.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.4 | 7.0 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.4 | 4.6 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.4 | 5.7 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.3 | 6.7 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.3 | 3.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.3 | 4.6 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.3 | 4.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.3 | 1.6 | GO:0002135 | CTP binding(GO:0002135) |
| 0.2 | 4.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 2.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 4.8 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.2 | 0.7 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 0.7 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) |
| 0.2 | 3.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 1.3 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.2 | 0.8 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.2 | 1.9 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.2 | 2.9 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.2 | 1.0 | GO:0070728 | leucine binding(GO:0070728) |
| 0.2 | 4.8 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.2 | 3.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 6.0 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.2 | 2.5 | GO:0010181 | FMN binding(GO:0010181) |
| 0.2 | 1.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.2 | 6.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.2 | 1.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 2.1 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.1 | 12.2 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.1 | 0.7 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.8 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 0.5 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 1.0 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 1.4 | GO:0048038 | quinone binding(GO:0048038) |
| 0.1 | 0.5 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 1.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.5 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.8 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 3.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.5 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.1 | 4.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 1.1 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 1.9 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 1.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 3.5 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 6.3 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 2.7 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 1.9 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.1 | 0.2 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.1 | 1.6 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 1.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.7 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 0.5 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 5.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 3.2 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 0.3 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 3.3 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 1.2 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 0.6 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 1.5 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.1 | 2.6 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 1.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 1.3 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 12.0 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 6.1 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.0 | 1.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.5 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 1.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.8 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.9 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 6.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 2.2 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 1.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.6 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.4 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.2 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 2.3 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 1.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.0 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 2.0 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 4.9 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.9 | GO:0005178 | integrin binding(GO:0005178) |