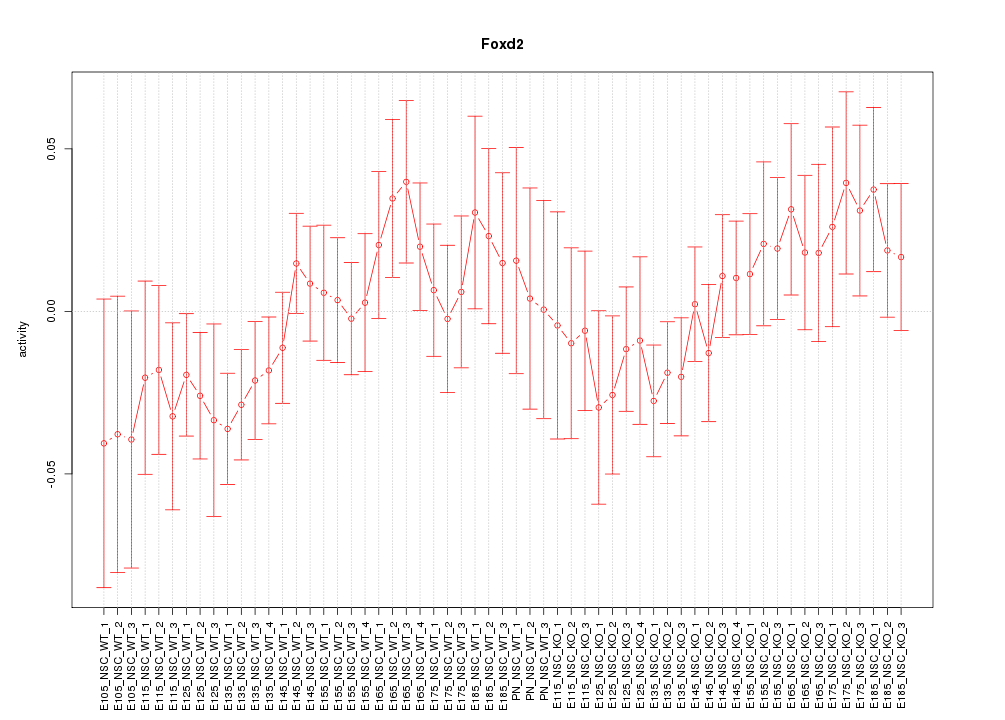
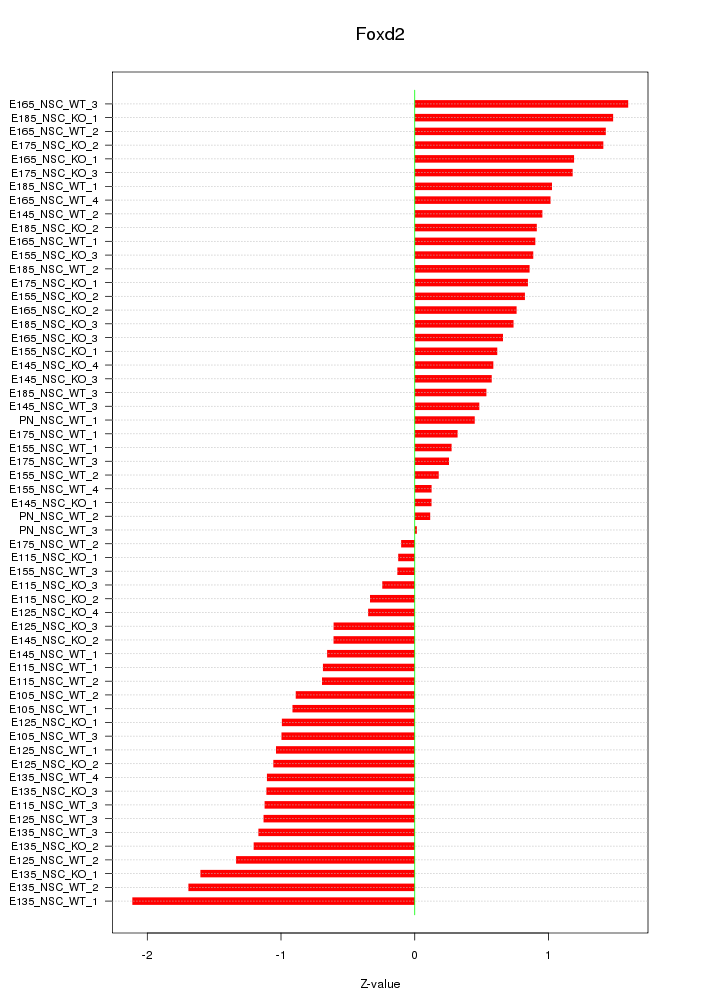
Motif ID: Foxd2
Z-value: 0.926
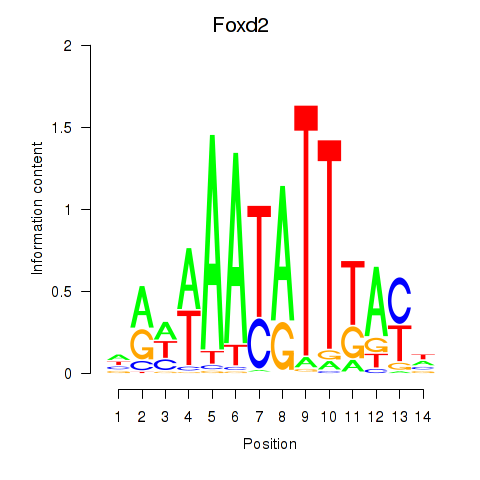
Transcription factors associated with Foxd2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Foxd2 | ENSMUSG00000055210.3 | Foxd2 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 13.1 | GO:0097393 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 2.1 | 6.3 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.8 | 5.1 | GO:1903056 | regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.7 | 5.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.7 | 5.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.6 | 2.6 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.6 | 3.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.5 | 2.7 | GO:0042636 | negative regulation of hair cycle(GO:0042636) progesterone secretion(GO:0042701) negative regulation of hair follicle development(GO:0051799) positive regulation of ovulation(GO:0060279) |
| 0.5 | 1.0 | GO:0032701 | negative regulation of interleukin-18 production(GO:0032701) |
| 0.4 | 3.7 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.4 | 4.0 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.4 | 2.1 | GO:0021814 | cell motility involved in cerebral cortex radial glia guided migration(GO:0021814) |
| 0.4 | 2.5 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.3 | 1.3 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.3 | 5.5 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.3 | 1.9 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.3 | 2.7 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.3 | 9.7 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.3 | 2.2 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.3 | 0.8 | GO:1901731 | calcium-mediated signaling using extracellular calcium source(GO:0035585) positive regulation of platelet aggregation(GO:1901731) |
| 0.2 | 2.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.2 | 3.4 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.2 | 1.5 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.2 | 0.8 | GO:1904721 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.2 | 2.0 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 | 3.1 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.1 | 0.9 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.8 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.1 | 4.2 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.4 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.2 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.1 | 2.5 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 4.0 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.1 | 0.2 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 1.0 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 5.7 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.1 | 1.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 2.6 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.0 | 0.4 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 2.1 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 0.4 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 1.0 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.3 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 2.4 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 7.4 | GO:0016337 | single organismal cell-cell adhesion(GO:0016337) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 13.1 | GO:1990421 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.9 | 2.7 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.6 | 6.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.3 | 4.2 | GO:0042581 | specific granule(GO:0042581) |
| 0.2 | 0.8 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.2 | 3.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 2.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 12.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 0.4 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 2.1 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 2.6 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 2.7 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 9.7 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 0.5 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 2.6 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 4.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 12.7 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 1.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 2.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.0 | 3.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 6.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 3.4 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 3.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.2 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 1.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.9 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.9 | GO:0030141 | secretory granule(GO:0030141) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 13.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 1.7 | 6.9 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 1.0 | 6.3 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.5 | 13.3 | GO:0042805 | actinin binding(GO:0042805) |
| 0.5 | 5.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.5 | 2.7 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.4 | 2.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.3 | 1.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.3 | 2.6 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.3 | 1.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 2.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.2 | 6.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 3.4 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 0.8 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.1 | 5.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 1.9 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 2.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.5 | GO:0001639 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) |
| 0.1 | 5.2 | GO:0018602 | sulfonate dioxygenase activity(GO:0000907) 2,4-dichlorophenoxyacetate alpha-ketoglutarate dioxygenase activity(GO:0018602) hypophosphite dioxygenase activity(GO:0034792) gibberellin 2-beta-dioxygenase activity(GO:0045543) C-19 gibberellin 2-beta-dioxygenase activity(GO:0052634) C-20 gibberellin 2-beta-dioxygenase activity(GO:0052635) |
| 0.1 | 2.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.9 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 4.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 1.0 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 3.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 9.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 0.8 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.4 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) |
| 0.0 | 2.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 12.8 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 3.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 3.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 2.5 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.2 | GO:0034951 | pivalyl-CoA mutase activity(GO:0034784) o-hydroxylaminobenzoate mutase activity(GO:0034951) lupeol synthase activity(GO:0042299) beta-amyrin synthase activity(GO:0042300) baruol synthase activity(GO:0080011) |
| 0.0 | 2.5 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |