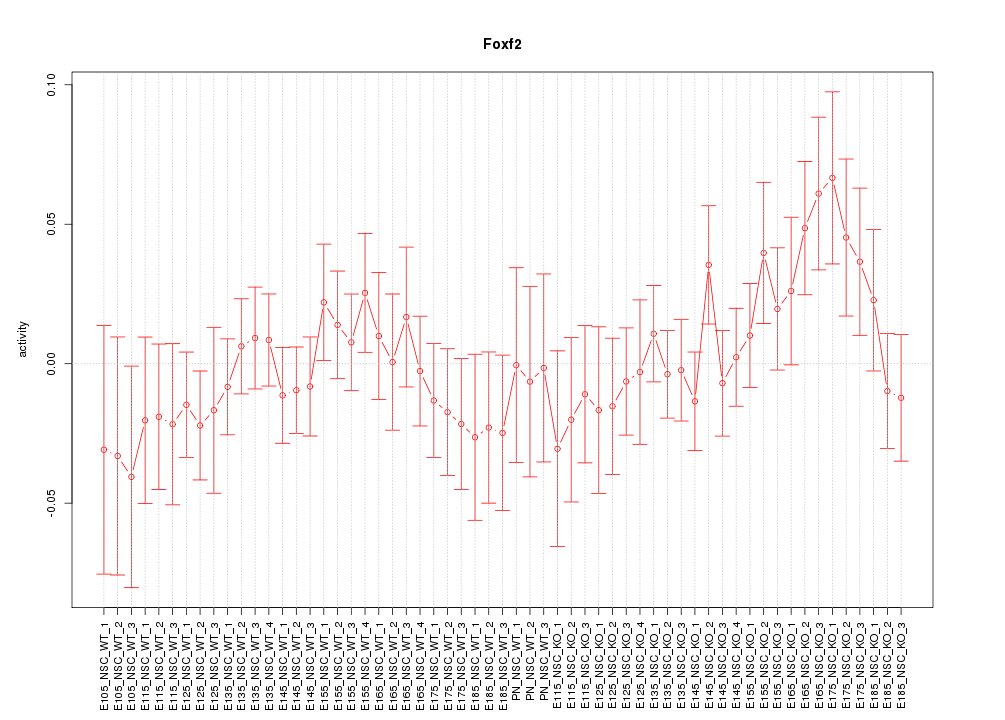
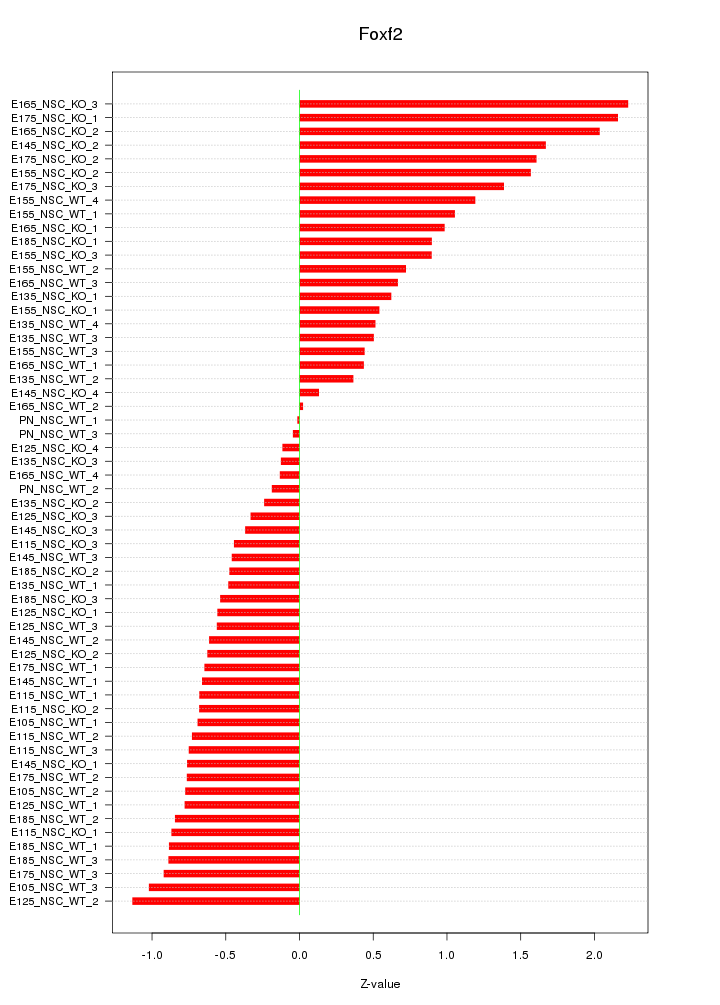
Motif ID: Foxf2
Z-value: 0.885
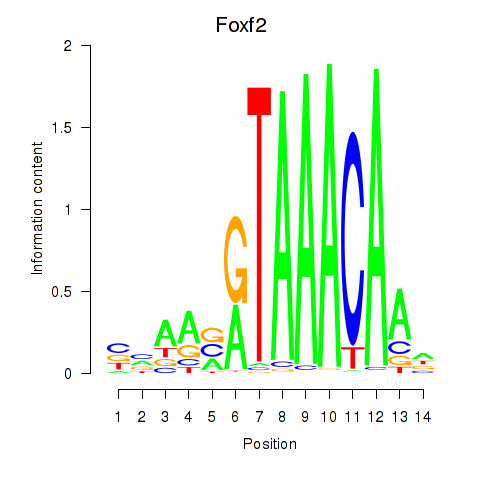
Transcription factors associated with Foxf2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Foxf2 | ENSMUSG00000038402.2 | Foxf2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxf2 | mm10_v2_chr13_+_31625802_31625816 | -0.03 | 8.0e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 15.8 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 2.0 | 20.1 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 1.5 | 4.4 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 1.1 | 3.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 1.1 | 6.4 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 1.1 | 14.8 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.8 | 6.4 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.8 | 13.6 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.7 | 2.6 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.6 | 6.1 | GO:0007379 | segment specification(GO:0007379) |
| 0.6 | 1.8 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.6 | 1.7 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.5 | 4.6 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.5 | 6.5 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.5 | 1.9 | GO:1903898 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.4 | 2.2 | GO:0070305 | response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.4 | 2.6 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.4 | 6.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.4 | 1.8 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.3 | 1.5 | GO:0071476 | cellular hypotonic response(GO:0071476) |
| 0.3 | 1.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.3 | 1.8 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.3 | 1.3 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.2 | 2.0 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.2 | 2.1 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.2 | 1.5 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.2 | 3.2 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.2 | 4.1 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.2 | 4.0 | GO:0010574 | regulation of vascular endothelial growth factor production(GO:0010574) |
| 0.2 | 1.6 | GO:0032693 | negative regulation of interleukin-10 production(GO:0032693) |
| 0.2 | 0.7 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.2 | 2.9 | GO:0071624 | positive regulation of granulocyte chemotaxis(GO:0071624) positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.2 | 1.4 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.1 | 0.7 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.1 | 10.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 1.7 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.7 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 0.5 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 | 1.5 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.1 | 1.0 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 3.9 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.1 | 3.7 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 1.5 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 0.6 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 2.9 | GO:0051926 | negative regulation of calcium ion transport(GO:0051926) |
| 0.1 | 0.8 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 3.6 | GO:0050729 | positive regulation of inflammatory response(GO:0050729) |
| 0.1 | 1.3 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 | 0.2 | GO:0009597 | detection of virus(GO:0009597) positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902237) |
| 0.0 | 0.6 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.3 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 3.2 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 11.5 | GO:0006396 | RNA processing(GO:0006396) |
| 0.0 | 2.4 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 11.5 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.8 | 26.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.7 | 2.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.6 | 4.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.5 | 1.5 | GO:0044754 | amphisome(GO:0044753) autolysosome(GO:0044754) |
| 0.4 | 1.5 | GO:0031673 | H zone(GO:0031673) |
| 0.3 | 3.2 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.2 | 0.7 | GO:0005712 | chiasma(GO:0005712) |
| 0.1 | 6.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 6.1 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 2.2 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.1 | 30.8 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.1 | 2.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 3.9 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 2.0 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.3 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 5.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 4.6 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 3.8 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.4 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.9 | 4.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.7 | 2.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.5 | 2.9 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.5 | 23.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.5 | 1.4 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.4 | 2.2 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.4 | 2.9 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.4 | 1.8 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.3 | 1.5 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.2 | 1.0 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.2 | 0.7 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 0.7 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 15.8 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 1.6 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 0.3 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.1 | 2.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 1.2 | GO:0070636 | single-strand selective uracil DNA N-glycosylase activity(GO:0017065) nicotinamide riboside hydrolase activity(GO:0070635) nicotinic acid riboside hydrolase activity(GO:0070636) deoxyribonucleoside 5'-monophosphate N-glycosidase activity(GO:0070694) |
| 0.1 | 4.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 1.5 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 1.8 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 1.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 2.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 7.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 2.1 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 1.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.6 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 3.9 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 2.0 | GO:0043851 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) rRNA (uridine-2'-O-)-methyltransferase activity(GO:0008650) rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) selenocysteine methyltransferase activity(GO:0016205) rRNA (adenine) methyltransferase activity(GO:0016433) rRNA (cytosine) methyltransferase activity(GO:0016434) rRNA (guanine) methyltransferase activity(GO:0016435) 1-phenanthrol methyltransferase activity(GO:0018707) protein-arginine N5-methyltransferase activity(GO:0019702) dimethylarsinite methyltransferase activity(GO:0034541) 4,5-dihydroxybenzo(a)pyrene methyltransferase activity(GO:0034807) 1-hydroxypyrene methyltransferase activity(GO:0034931) 1-hydroxy-6-methoxypyrene methyltransferase activity(GO:0034933) demethylmenaquinone methyltransferase activity(GO:0043770) cobalt-precorrin-6B C5-methyltransferase activity(GO:0043776) cobalt-precorrin-7 C15-methyltransferase activity(GO:0043777) cobalt-precorrin-5B C1-methyltransferase activity(GO:0043780) cobalt-precorrin-3 C17-methyltransferase activity(GO:0043782) dimethylamine methyltransferase activity(GO:0043791) hydroxyneurosporene-O-methyltransferase activity(GO:0043803) tRNA (adenine-57, 58-N(1)-) methyltransferase activity(GO:0043827) methylamine-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043833) trimethylamine methyltransferase activity(GO:0043834) methanol-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043851) monomethylamine methyltransferase activity(GO:0043852) P-methyltransferase activity(GO:0051994) Se-methyltransferase activity(GO:0051995) 2-phytyl-1,4-naphthoquinone methyltransferase activity(GO:0052624) tRNA (uracil-2'-O-)-methyltransferase activity(GO:0052665) tRNA (cytosine-2'-O-)-methyltransferase activity(GO:0052666) phosphomethylethanolamine N-methyltransferase activity(GO:0052667) tRNA (cytosine-3-)-methyltransferase activity(GO:0052735) rRNA (cytosine-2'-O-)-methyltransferase activity(GO:0070677) rRNA (cytosine-N4-)-methyltransferase activity(GO:0071424) trihydroxyferuloyl spermidine O-methyltransferase activity(GO:0080012) |
| 0.0 | 5.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.2 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 6.6 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.9 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.0 | 0.8 | GO:0035496 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) O antigen polymerase activity(GO:0008755) lipopolysaccharide-1,6-galactosyltransferase activity(GO:0008921) cellulose synthase activity(GO:0016759) 9-phenanthrol UDP-glucuronosyltransferase activity(GO:0018715) 1-phenanthrol glycosyltransferase activity(GO:0018716) 9-phenanthrol glycosyltransferase activity(GO:0018717) 1,2-dihydroxy-phenanthrene glycosyltransferase activity(GO:0018718) phenanthrol glycosyltransferase activity(GO:0019112) alpha-1,2-galactosyltransferase activity(GO:0031278) dolichyl pyrophosphate Man7GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0033556) endogalactosaminidase activity(GO:0033931) lipopolysaccharide-1,5-galactosyltransferase activity(GO:0035496) dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0042283) inositol phosphoceramide synthase activity(GO:0045140) alpha-(1->3)-fucosyltransferase activity(GO:0046920) indole-3-butyrate beta-glucosyltransferase activity(GO:0052638) salicylic acid glucosyltransferase (ester-forming) activity(GO:0052639) salicylic acid glucosyltransferase (glucoside-forming) activity(GO:0052640) benzoic acid glucosyltransferase activity(GO:0052641) chondroitin hydrolase activity(GO:0052757) dolichyl-pyrophosphate Man7GlcNAc2 alpha-1,6-mannosyltransferase activity(GO:0052824) cytokinin 9-beta-glucosyltransferase activity(GO:0080062) |
| 0.0 | 0.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 1.5 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 0.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |