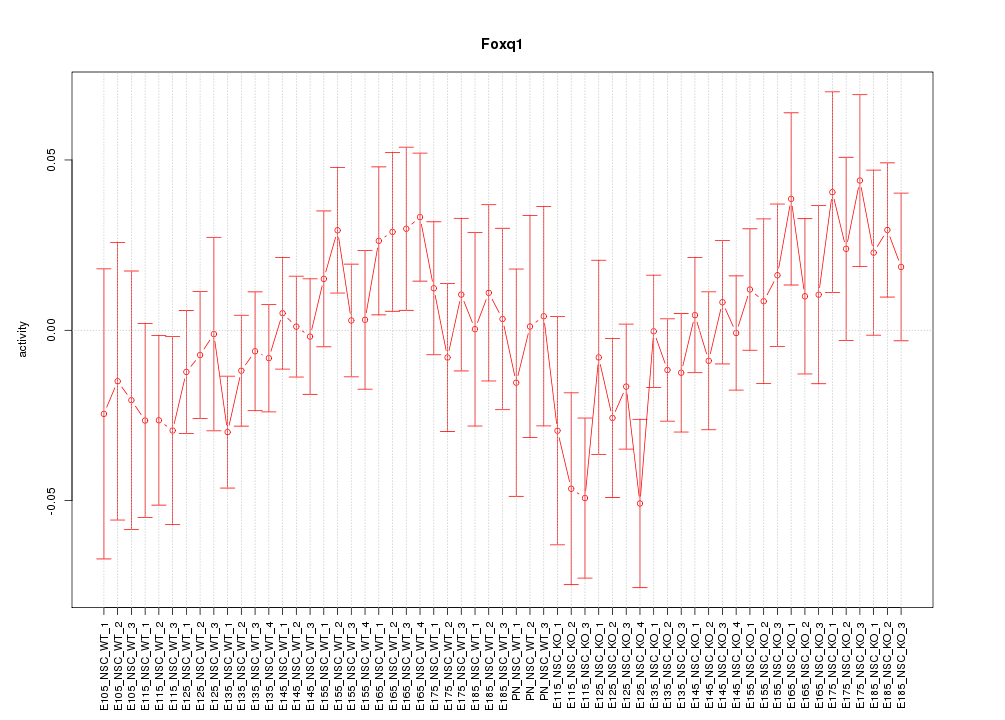
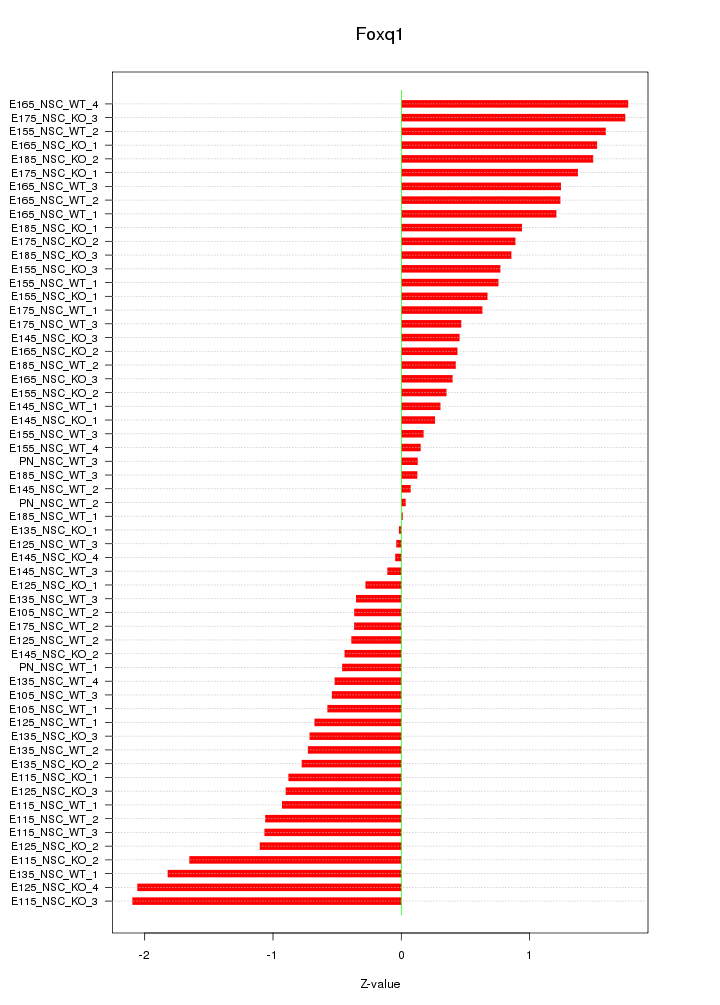
Motif ID: Foxq1
Z-value: 0.922
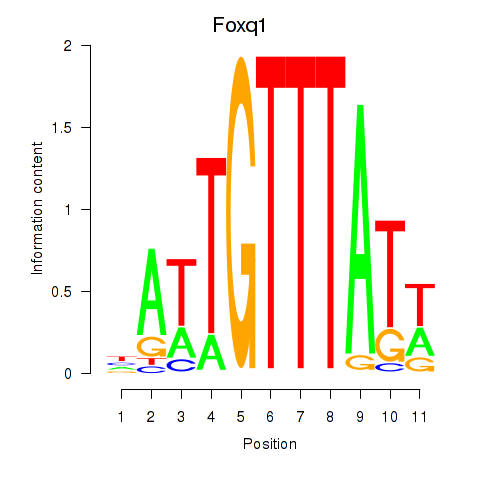
Transcription factors associated with Foxq1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Foxq1 | ENSMUSG00000038415.8 | Foxq1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxq1 | mm10_v2_chr13_+_31558157_31558176 | 0.38 | 2.9e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 9.8 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 3.0 | 9.1 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 2.2 | 4.5 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 1.4 | 4.2 | GO:0032650 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) interleukin-1 alpha secretion(GO:0050703) |
| 1.0 | 2.9 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.8 | 3.3 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.8 | 8.2 | GO:0007379 | segment specification(GO:0007379) |
| 0.7 | 5.3 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.6 | 6.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.6 | 1.8 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.5 | 7.3 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.5 | 3.5 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.5 | 3.3 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.4 | 3.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.3 | 9.7 | GO:0095500 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.3 | 1.6 | GO:0090164 | asymmetric Golgi ribbon formation(GO:0090164) |
| 0.3 | 10.5 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.3 | 1.4 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.3 | 5.5 | GO:0007614 | short-term memory(GO:0007614) |
| 0.2 | 1.2 | GO:0071476 | cellular hypotonic response(GO:0071476) |
| 0.2 | 2.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 1.7 | GO:0006537 | glutamate biosynthetic process(GO:0006537) proline biosynthetic process(GO:0006561) |
| 0.2 | 0.6 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.2 | 2.3 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.2 | 4.1 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.2 | 1.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.2 | 3.4 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.2 | 1.8 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 3.6 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.1 | 2.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 1.2 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 1.9 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.6 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.1 | 1.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 6.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.6 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 3.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 3.2 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 3.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 6.5 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 0.4 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 0.5 | GO:1901216 | positive regulation of neuron death(GO:1901216) |
| 0.0 | 0.3 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.2 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.0 | 1.5 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 4.3 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.4 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 1.8 | GO:0055072 | iron ion homeostasis(GO:0055072) |
| 0.0 | 0.2 | GO:0002643 | regulation of tolerance induction(GO:0002643) |
| 0.0 | 3.6 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 6.1 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.2 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.0 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 3.1 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 1.3 | GO:0098792 | xenophagy(GO:0098792) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 9.8 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.9 | 9.1 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.8 | 3.3 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.3 | 1.8 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.3 | 4.2 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.3 | 2.9 | GO:0000805 | X chromosome(GO:0000805) |
| 0.2 | 8.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.2 | 2.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 4.8 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 8.2 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 0.7 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 0.6 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 1.8 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.6 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 0.3 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 4.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 0.7 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 4.1 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 3.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.7 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 2.3 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 4.5 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 10.0 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 1.6 | 9.8 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 1.6 | 6.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 1.2 | 3.6 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.8 | 5.3 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.7 | 4.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.7 | 3.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.4 | 2.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.4 | 3.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.3 | 1.8 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.3 | 1.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.2 | 1.5 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.2 | 2.8 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.2 | 1.2 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.2 | 2.6 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 2.5 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.1 | 1.4 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 0.7 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 3.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 16.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 1.9 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 0.9 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 2.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 1.7 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 3.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.3 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.6 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 1.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.4 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 1.2 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 1.6 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 14.2 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 5.6 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 2.9 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 3.0 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.0 | 3.0 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.6 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.8 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 7.1 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.4 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.8 | GO:0002039 | p53 binding(GO:0002039) |