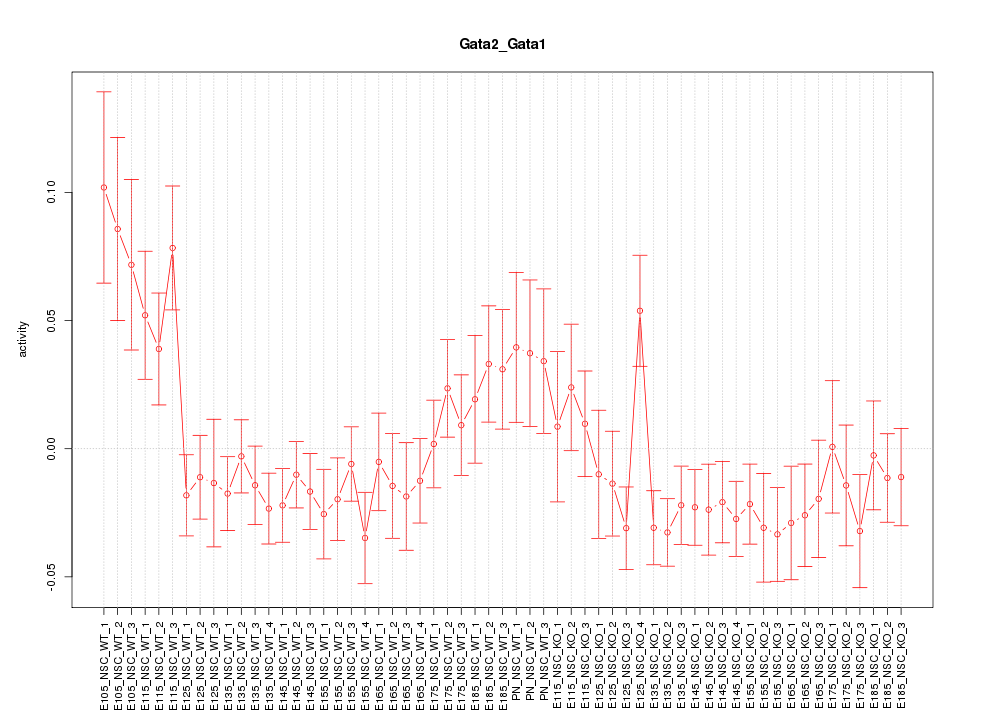
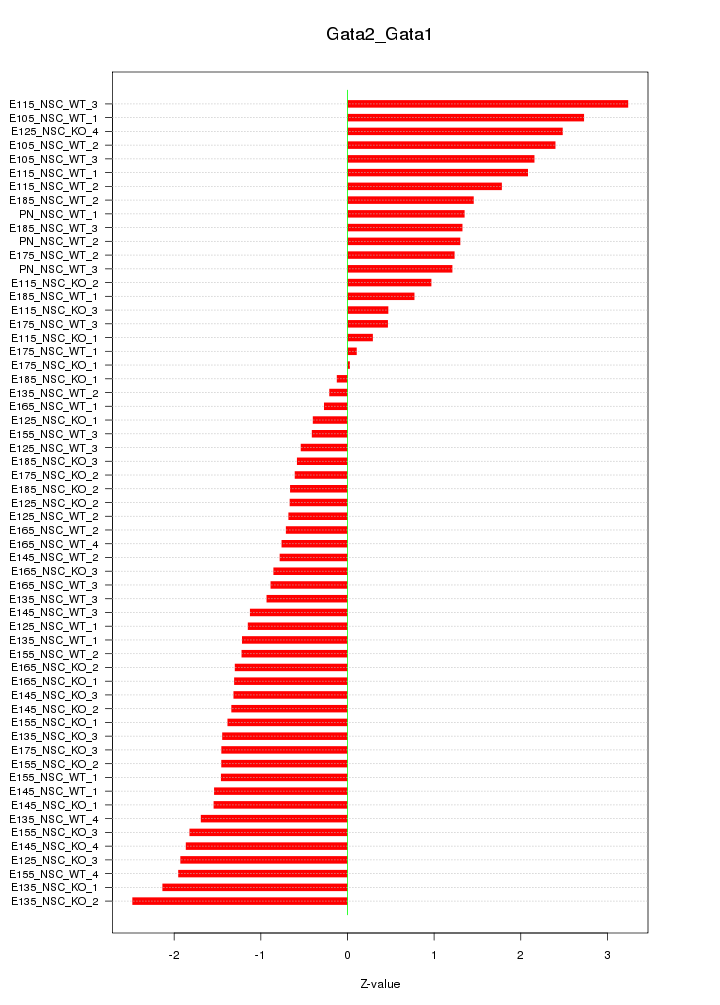
Motif ID: Gata2_Gata1
Z-value: 1.408

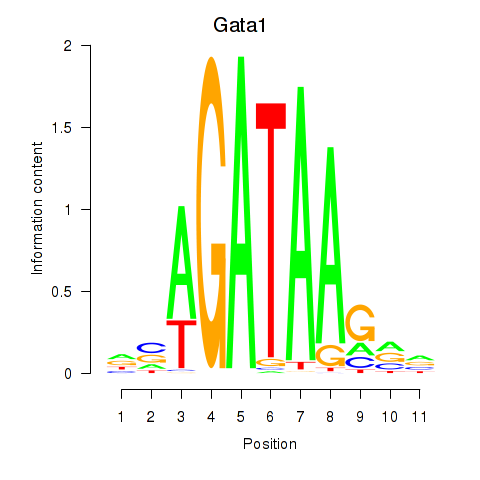
Transcription factors associated with Gata2_Gata1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Gata1 | ENSMUSG00000031162.8 | Gata1 |
| Gata2 | ENSMUSG00000015053.8 | Gata2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gata1 | mm10_v2_chrX_-_7967817_7967910 | 0.78 | 3.9e-13 | Click! |
| Gata2 | mm10_v2_chr6_+_88198656_88198675 | 0.38 | 3.1e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 19.8 | 98.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 6.1 | 18.3 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 3.4 | 10.3 | GO:0032650 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) cardiac cell fate determination(GO:0060913) |
| 3.4 | 10.2 | GO:0030300 | regulation of intestinal cholesterol absorption(GO:0030300) |
| 2.8 | 16.6 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 2.5 | 25.2 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 2.4 | 16.8 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 2.1 | 12.5 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 1.9 | 9.3 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 1.8 | 16.4 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 1.8 | 12.7 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 1.5 | 4.6 | GO:0015920 | regulation of phosphatidylcholine catabolic process(GO:0010899) lipopolysaccharide transport(GO:0015920) |
| 1.5 | 6.0 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 1.4 | 14.1 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 1.4 | 5.4 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 1.3 | 8.0 | GO:0002317 | plasma cell differentiation(GO:0002317) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 1.3 | 6.5 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) positive regulation of blood vessel remodeling(GO:2000504) |
| 1.3 | 2.6 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 1.2 | 4.7 | GO:0097168 | condensed mesenchymal cell proliferation(GO:0072137) mesenchymal stem cell proliferation(GO:0097168) |
| 1.1 | 6.3 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 1.0 | 9.5 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.9 | 4.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.8 | 8.5 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.8 | 10.8 | GO:0046697 | decidualization(GO:0046697) |
| 0.8 | 3.8 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.7 | 11.0 | GO:0034377 | plasma lipoprotein particle assembly(GO:0034377) |
| 0.7 | 2.2 | GO:0048352 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) |
| 0.6 | 2.5 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.6 | 1.8 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.6 | 10.3 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.6 | 14.7 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.6 | 1.8 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.6 | 1.7 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.6 | 2.2 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.5 | 5.4 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.5 | 3.1 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.5 | 12.0 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.4 | 4.8 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.4 | 1.2 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.4 | 1.2 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.4 | 5.5 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.4 | 3.8 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.3 | 2.6 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.3 | 2.1 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.3 | 20.7 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.3 | 4.9 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.2 | 1.7 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.2 | 4.6 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.2 | 1.3 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.2 | 0.9 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.2 | 1.6 | GO:0046036 | GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.1 | 1.3 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 0.6 | GO:1904995 | negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.1 | 2.2 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 | 3.7 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.1 | 3.7 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 3.1 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 | 11.4 | GO:0030168 | platelet activation(GO:0030168) |
| 0.1 | 6.4 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.1 | 1.3 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 | 1.5 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.1 | 1.2 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 2.2 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.1 | 6.3 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 2.0 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.1 | 0.5 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 5.4 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.0 | 0.7 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 5.6 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.2 | GO:1901162 | tryptophan metabolic process(GO:0006568) indolalkylamine metabolic process(GO:0006586) serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.3 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.3 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 1.5 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.9 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.1 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.3 | 98.8 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 3.4 | 20.7 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 3.3 | 16.6 | GO:0005861 | troponin complex(GO:0005861) |
| 1.3 | 6.4 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 1.0 | 10.2 | GO:0098533 | ATPase dependent transmembrane transport complex(GO:0098533) |
| 0.7 | 9.3 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.5 | 8.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.5 | 5.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.3 | 1.3 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.3 | 5.1 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.2 | 0.7 | GO:1990393 | 3M complex(GO:1990393) |
| 0.2 | 2.2 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.2 | 6.3 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 2.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 2.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 3.9 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 2.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 7.9 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 0.9 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.5 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 9.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 4.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 8.2 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.1 | 14.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.7 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 2.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.1 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 2.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 8.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 4.7 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.2 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 6.7 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 3.8 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.9 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 10.3 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 1.2 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 33.3 | GO:0005576 | extracellular region(GO:0005576) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 19.8 | 98.8 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 5.5 | 16.6 | GO:0030172 | troponin C binding(GO:0030172) |
| 5.4 | 16.2 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 4.2 | 12.5 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 2.0 | 7.8 | GO:0004921 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 1.8 | 5.5 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 1.6 | 9.5 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 1.5 | 4.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 1.5 | 4.6 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 1.1 | 11.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 1.1 | 5.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 1.1 | 6.3 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 1.0 | 5.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.9 | 25.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.9 | 14.0 | GO:0016594 | glycine binding(GO:0016594) |
| 0.8 | 18.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.8 | 4.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.7 | 2.2 | GO:0005119 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) hedgehog family protein binding(GO:0097108) |
| 0.7 | 6.4 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.7 | 6.4 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.7 | 2.1 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.6 | 9.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.6 | 5.4 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.6 | 1.8 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) arginine binding(GO:0034618) |
| 0.6 | 1.7 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.6 | 10.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.5 | 6.0 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.5 | 3.8 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.4 | 5.4 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.4 | 2.6 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.4 | 2.2 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.4 | 4.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.3 | 15.1 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.3 | 5.7 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.3 | 5.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.3 | 6.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.2 | 8.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 8.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.2 | 1.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.2 | 5.6 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.1 | 2.6 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 0.6 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.1 | 12.0 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.1 | 3.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 11.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 0.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.5 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 0.2 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.1 | 1.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 3.0 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 2.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 3.8 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 3.7 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.1 | 0.2 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 13.5 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 3.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.9 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.2 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 2.5 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 2.4 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.8 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 1.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 12.5 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 4.8 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 21.0 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 3.7 | GO:0005096 | GTPase activator activity(GO:0005096) |