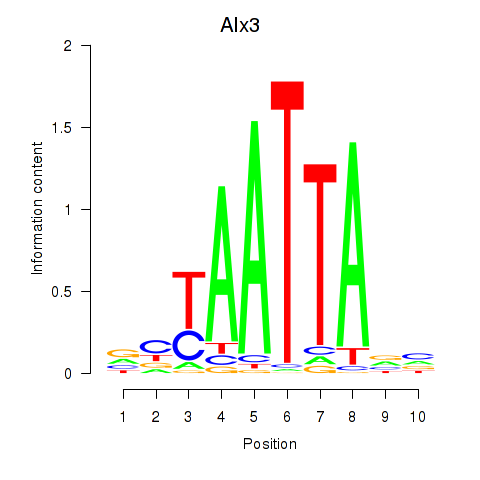
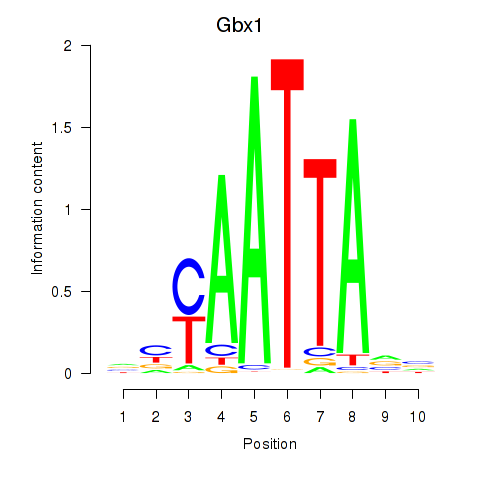
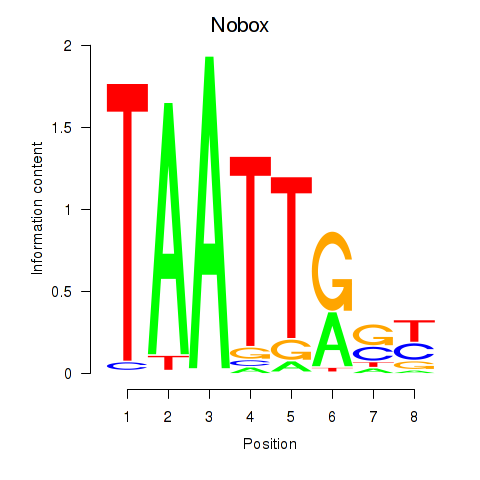
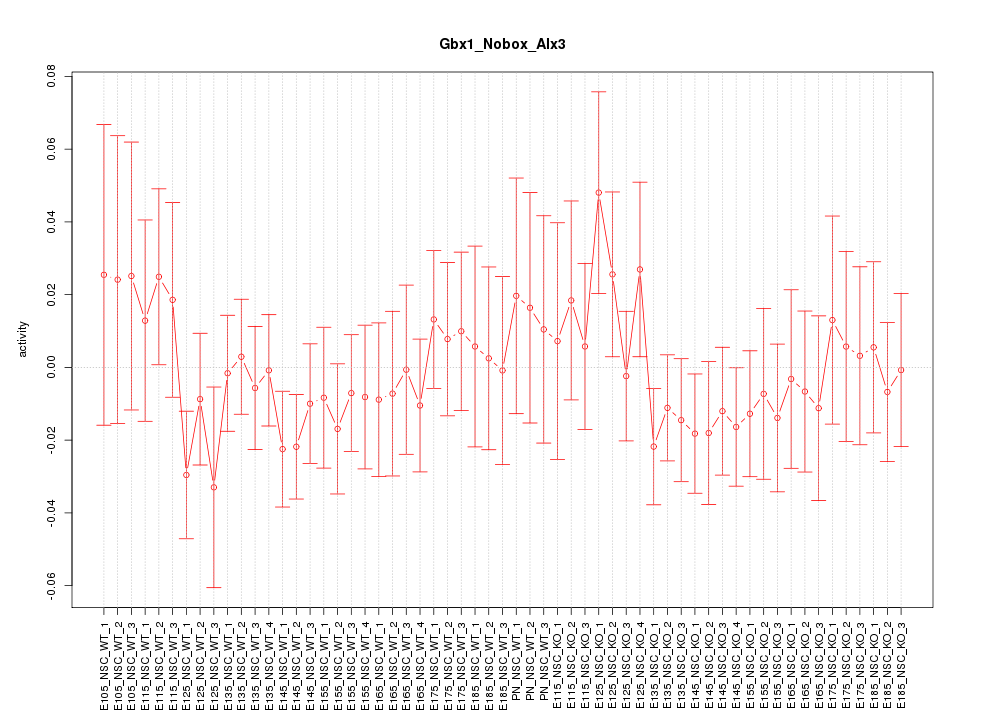
| 2.7 |
8.1 |
GO:0072204 |
cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 2.0 |
6.1 |
GO:0090425 |
hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 1.2 |
9.2 |
GO:0035726 |
common myeloid progenitor cell proliferation(GO:0035726) |
| 1.1 |
4.3 |
GO:0046552 |
eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 1.0 |
3.0 |
GO:0048818 |
embryonic nail plate morphogenesis(GO:0035880) positive regulation of hair follicle maturation(GO:0048818) positive regulation of catagen(GO:0051795) frontal suture morphogenesis(GO:0060364) |
| 1.0 |
2.9 |
GO:0021759 |
globus pallidus development(GO:0021759) |
| 0.8 |
2.5 |
GO:0060743 |
epithelial cell maturation involved in prostate gland development(GO:0060743) alveolar secondary septum development(GO:0061144) |
| 0.8 |
6.2 |
GO:0033353 |
S-adenosylmethionine cycle(GO:0033353) |
| 0.7 |
1.4 |
GO:0072106 |
regulation of ureteric bud formation(GO:0072106) positive regulation of ureteric bud formation(GO:0072107) |
| 0.7 |
4.1 |
GO:0006123 |
mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.6 |
11.6 |
GO:0001833 |
inner cell mass cell proliferation(GO:0001833) |
| 0.6 |
1.8 |
GO:0003278 |
apoptotic process involved in heart morphogenesis(GO:0003278) regulation of vascular wound healing(GO:0061043) glomerular mesangial cell differentiation(GO:0072008) glomerular mesangial cell development(GO:0072144) |
| 0.6 |
7.2 |
GO:0031017 |
exocrine pancreas development(GO:0031017) |
| 0.5 |
2.0 |
GO:0051798 |
positive regulation of hair follicle development(GO:0051798) |
| 0.5 |
1.5 |
GO:0042663 |
regulation of endodermal cell fate specification(GO:0042663) |
| 0.5 |
1.9 |
GO:0001550 |
ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.5 |
1.4 |
GO:2000977 |
regulation of forebrain neuron differentiation(GO:2000977) |
| 0.5 |
2.8 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.4 |
1.8 |
GO:0000707 |
meiotic DNA recombinase assembly(GO:0000707) resolution of recombination intermediates(GO:0071139) |
| 0.4 |
3.1 |
GO:0060872 |
semicircular canal morphogenesis(GO:0048752) semicircular canal development(GO:0060872) |
| 0.4 |
2.4 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.4 |
12.4 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.4 |
1.2 |
GO:0008228 |
opsonization(GO:0008228) modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.4 |
1.1 |
GO:1902608 |
testosterone biosynthetic process(GO:0061370) regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.3 |
1.0 |
GO:0060166 |
olfactory pit development(GO:0060166) |
| 0.3 |
1.0 |
GO:0030862 |
neuroblast division in subventricular zone(GO:0021849) regulation of polarized epithelial cell differentiation(GO:0030860) positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.3 |
1.0 |
GO:0090271 |
positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.3 |
1.0 |
GO:0003162 |
atrioventricular node development(GO:0003162) |
| 0.3 |
0.3 |
GO:0032472 |
Golgi calcium ion transport(GO:0032472) |
| 0.3 |
1.9 |
GO:0042795 |
snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.3 |
1.5 |
GO:0006348 |
chromatin silencing at telomere(GO:0006348) |
| 0.3 |
2.6 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.3 |
1.1 |
GO:0090324 |
negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.3 |
1.1 |
GO:0035553 |
oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.3 |
1.6 |
GO:1902261 |
positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.3 |
1.0 |
GO:0006742 |
NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.2 |
2.2 |
GO:0045602 |
negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.2 |
1.0 |
GO:0010700 |
negative regulation of norepinephrine secretion(GO:0010700) |
| 0.2 |
1.6 |
GO:0015808 |
L-alanine transport(GO:0015808) |
| 0.2 |
2.9 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.2 |
0.7 |
GO:2000851 |
positive regulation of glucocorticoid secretion(GO:2000851) regulation of corticosterone secretion(GO:2000852) |
| 0.2 |
0.9 |
GO:0071596 |
ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.2 |
1.3 |
GO:0070562 |
regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.2 |
2.3 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 |
2.3 |
GO:2000671 |
regulation of motor neuron apoptotic process(GO:2000671) |
| 0.2 |
1.2 |
GO:0010624 |
regulation of Schwann cell proliferation(GO:0010624) |
| 0.2 |
5.1 |
GO:0002089 |
lens morphogenesis in camera-type eye(GO:0002089) |
| 0.2 |
0.4 |
GO:0042536 |
negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.2 |
2.2 |
GO:0051482 |
positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.2 |
2.9 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 |
0.8 |
GO:0015692 |
vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.2 |
5.2 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.2 |
2.7 |
GO:0042541 |
hemoglobin biosynthetic process(GO:0042541) |
| 0.1 |
1.0 |
GO:0070447 |
positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.1 |
1.7 |
GO:0044458 |
motile cilium assembly(GO:0044458) |
| 0.1 |
0.4 |
GO:0001543 |
ovarian follicle rupture(GO:0001543) |
| 0.1 |
0.4 |
GO:0030327 |
prenylated protein catabolic process(GO:0030327) |
| 0.1 |
5.0 |
GO:0045494 |
photoreceptor cell maintenance(GO:0045494) |
| 0.1 |
1.0 |
GO:1990118 |
sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 |
0.8 |
GO:0052695 |
cellular glucuronidation(GO:0052695) |
| 0.1 |
3.7 |
GO:0006907 |
pinocytosis(GO:0006907) |
| 0.1 |
0.5 |
GO:0006538 |
glutamate catabolic process(GO:0006538) |
| 0.1 |
2.8 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.1 |
0.7 |
GO:0072488 |
ammonium transmembrane transport(GO:0072488) |
| 0.1 |
4.0 |
GO:0045879 |
negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.1 |
0.5 |
GO:1900740 |
regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 |
0.6 |
GO:0002317 |
plasma cell differentiation(GO:0002317) |
| 0.1 |
3.1 |
GO:0046677 |
response to antibiotic(GO:0046677) |
| 0.1 |
0.9 |
GO:0035095 |
behavioral response to nicotine(GO:0035095) |
| 0.1 |
0.6 |
GO:0021756 |
striatum development(GO:0021756) |
| 0.1 |
0.3 |
GO:0006419 |
alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 |
0.4 |
GO:0060690 |
epithelial cell differentiation involved in salivary gland development(GO:0060690) |
| 0.1 |
0.3 |
GO:1900275 |
negative regulation of phospholipase C activity(GO:1900275) |
| 0.1 |
0.9 |
GO:0015721 |
bile acid and bile salt transport(GO:0015721) |
| 0.1 |
1.1 |
GO:0006107 |
oxaloacetate metabolic process(GO:0006107) |
| 0.1 |
0.9 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 |
0.7 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 |
1.0 |
GO:0019985 |
translesion synthesis(GO:0019985) |
| 0.1 |
0.4 |
GO:0038018 |
Wnt receptor catabolic process(GO:0038018) |
| 0.1 |
0.8 |
GO:0035520 |
monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 |
0.4 |
GO:0034047 |
regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.1 |
0.1 |
GO:1900747 |
negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 |
2.2 |
GO:0031365 |
N-terminal protein amino acid modification(GO:0031365) |
| 0.1 |
0.3 |
GO:0051661 |
maintenance of centrosome location(GO:0051661) |
| 0.1 |
0.8 |
GO:0006268 |
DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 |
0.9 |
GO:0030497 |
fatty acid elongation(GO:0030497) |
| 0.1 |
3.1 |
GO:0046676 |
negative regulation of insulin secretion(GO:0046676) |
| 0.1 |
1.2 |
GO:0032291 |
central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 |
4.5 |
GO:0003073 |
regulation of systemic arterial blood pressure(GO:0003073) |
| 0.1 |
5.5 |
GO:0001889 |
liver development(GO:0001889) |
| 0.1 |
0.9 |
GO:0038092 |
nodal signaling pathway(GO:0038092) |
| 0.1 |
1.0 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 |
0.2 |
GO:0002677 |
negative regulation of chronic inflammatory response(GO:0002677) |
| 0.0 |
1.3 |
GO:0006101 |
tricarboxylic acid cycle(GO:0006099) citrate metabolic process(GO:0006101) |
| 0.0 |
0.5 |
GO:1903504 |
regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 |
0.5 |
GO:0071763 |
nuclear membrane organization(GO:0071763) |
| 0.0 |
0.2 |
GO:0010359 |
regulation of anion channel activity(GO:0010359) |
| 0.0 |
6.0 |
GO:0030178 |
negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 |
2.6 |
GO:0042475 |
odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 |
0.4 |
GO:0070475 |
rRNA base methylation(GO:0070475) |
| 0.0 |
3.6 |
GO:0072332 |
intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 |
0.2 |
GO:0060648 |
mammary gland bud morphogenesis(GO:0060648) |
| 0.0 |
0.1 |
GO:0006667 |
sphinganine metabolic process(GO:0006667) |
| 0.0 |
0.0 |
GO:0007223 |
Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 |
0.2 |
GO:0038128 |
ERBB2 signaling pathway(GO:0038128) |
| 0.0 |
0.4 |
GO:0097034 |
mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 |
0.4 |
GO:0071578 |
zinc II ion transmembrane import(GO:0071578) |
| 0.0 |
0.3 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 |
0.6 |
GO:0050860 |
negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 |
0.6 |
GO:0021702 |
cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 |
0.1 |
GO:2001271 |
negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 |
0.1 |
GO:0043922 |
negative regulation by host of viral transcription(GO:0043922) |
| 0.0 |
0.2 |
GO:0007195 |
adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 |
0.1 |
GO:1903977 |
positive regulation of Schwann cell migration(GO:1900149) positive regulation of glial cell migration(GO:1903977) regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.0 |
0.4 |
GO:0001886 |
endothelial cell morphogenesis(GO:0001886) |
| 0.0 |
0.3 |
GO:0036371 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 |
0.5 |
GO:0043552 |
positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 |
0.4 |
GO:0001553 |
luteinization(GO:0001553) |
| 0.0 |
1.0 |
GO:0051693 |
actin filament capping(GO:0051693) |
| 0.0 |
0.2 |
GO:0036233 |
glycine import(GO:0036233) |
| 0.0 |
0.3 |
GO:0046548 |
retinal rod cell development(GO:0046548) |
| 0.0 |
0.4 |
GO:0035335 |
peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 |
0.1 |
GO:0043415 |
positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 |
0.1 |
GO:0044791 |
modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 |
4.0 |
GO:0008360 |
regulation of cell shape(GO:0008360) |
| 0.0 |
0.2 |
GO:0045974 |
miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 |
0.2 |
GO:0035094 |
response to nicotine(GO:0035094) |
| 0.0 |
1.8 |
GO:0019882 |
antigen processing and presentation(GO:0019882) |
| 0.0 |
0.5 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 |
1.3 |
GO:0050680 |
negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.0 |
0.2 |
GO:0086016 |
AV node cell action potential(GO:0086016) AV node cell to bundle of His cell signaling(GO:0086027) AV node cell to bundle of His cell communication(GO:0086067) |
| 0.0 |
0.1 |
GO:0070584 |
mitochondrion morphogenesis(GO:0070584) |
| 0.0 |
0.4 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 |
0.7 |
GO:0007029 |
endoplasmic reticulum organization(GO:0007029) |
| 0.0 |
0.2 |
GO:0030150 |
protein import into mitochondrial matrix(GO:0030150) |
| 0.0 |
0.1 |
GO:0051764 |
actin crosslink formation(GO:0051764) |
| 0.0 |
0.9 |
GO:0050728 |
negative regulation of inflammatory response(GO:0050728) |
| 0.0 |
0.1 |
GO:0019348 |
dolichol-linked oligosaccharide biosynthetic process(GO:0006488) dolichol metabolic process(GO:0019348) |
| 0.0 |
0.3 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 |
0.3 |
GO:0030212 |
hyaluronan metabolic process(GO:0030212) |
| 0.0 |
0.3 |
GO:0006940 |
regulation of smooth muscle contraction(GO:0006940) |
| 0.0 |
0.1 |
GO:0090129 |
positive regulation of synapse maturation(GO:0090129) |
| 0.0 |
0.3 |
GO:0050819 |
negative regulation of coagulation(GO:0050819) |
| 0.0 |
0.3 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
0.1 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |