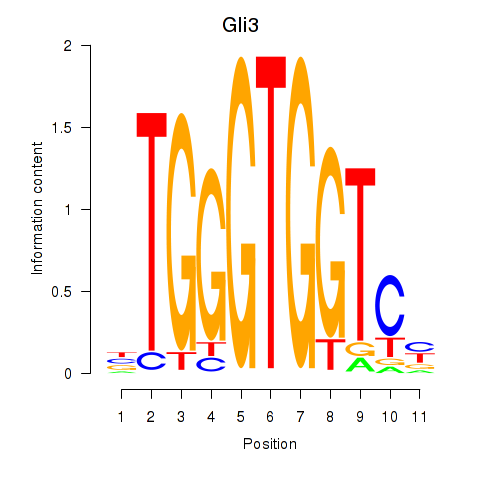
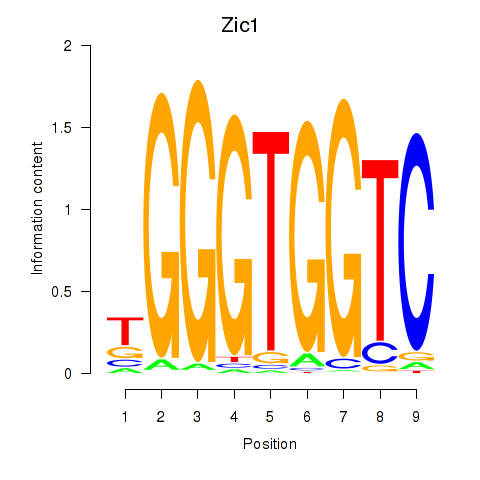
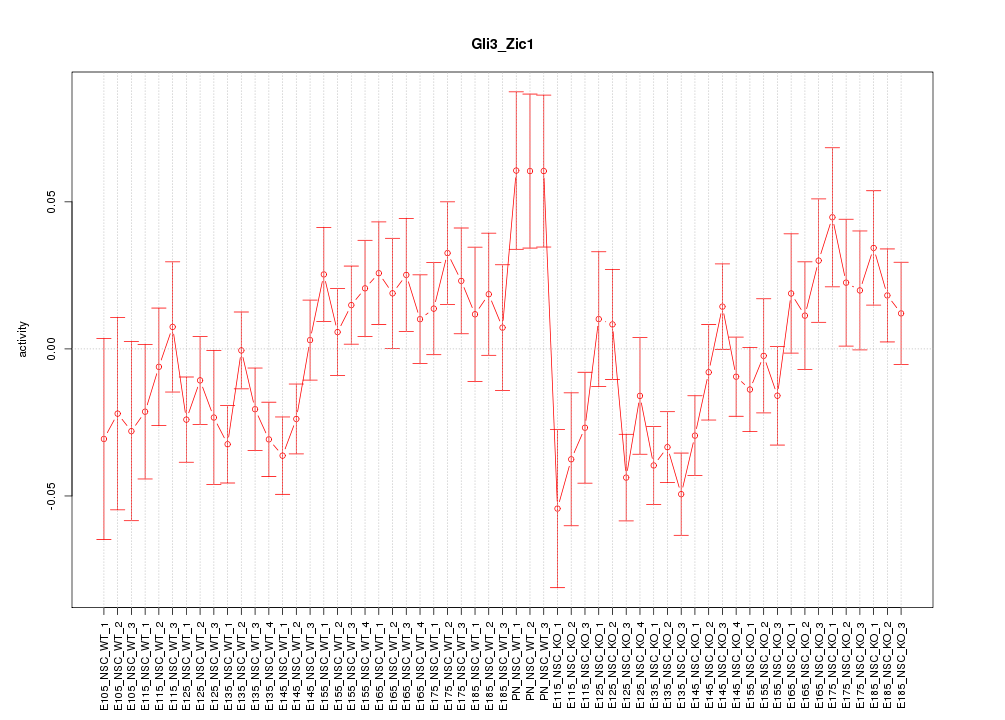
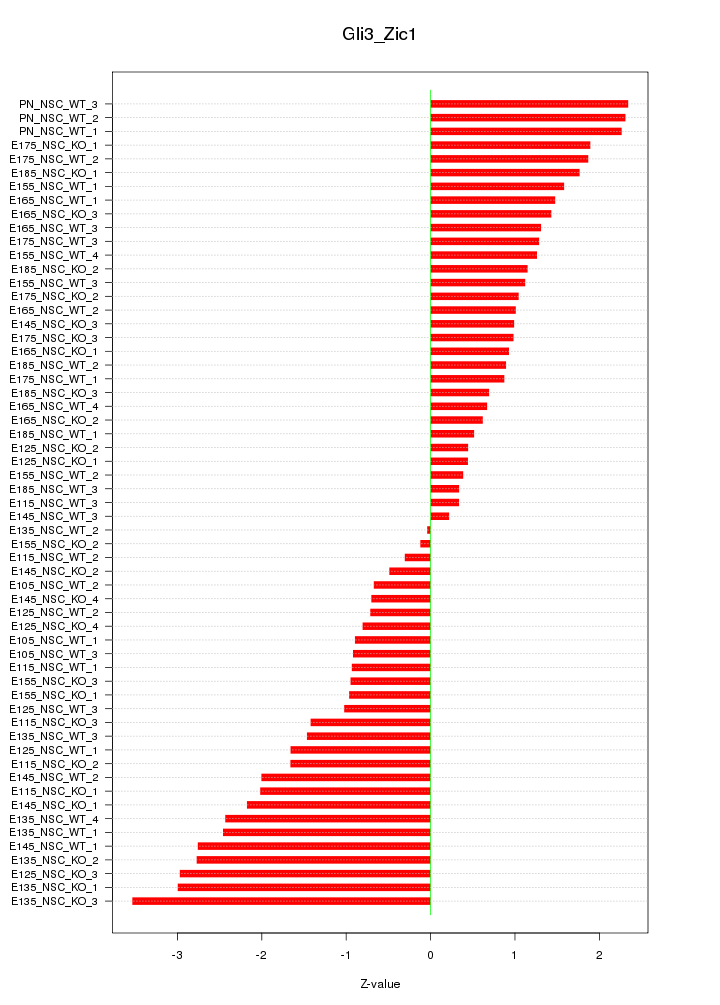
| 3.1 |
9.3 |
GO:0032241 |
positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 2.5 |
9.9 |
GO:0060032 |
notochord regression(GO:0060032) |
| 2.4 |
14.5 |
GO:0007021 |
tubulin complex assembly(GO:0007021) |
| 2.4 |
7.1 |
GO:0045763 |
regulation of arginine metabolic process(GO:0000821) negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 2.1 |
6.4 |
GO:2001139 |
negative regulation of postsynaptic membrane organization(GO:1901627) negative regulation of dendritic spine maintenance(GO:1902951) negative regulation of phospholipid efflux(GO:1902999) regulation of lipid transport across blood brain barrier(GO:1903000) negative regulation of lipid transport across blood brain barrier(GO:1903001) positive regulation of lipid transport across blood brain barrier(GO:1903002) negative regulation of phospholipid transport(GO:2001139) |
| 2.1 |
12.4 |
GO:0060024 |
rhythmic synaptic transmission(GO:0060024) |
| 1.9 |
5.6 |
GO:1903903 |
striated muscle atrophy(GO:0014891) positive regulation of keratinocyte apoptotic process(GO:1902174) regulation of establishment of T cell polarity(GO:1903903) |
| 1.8 |
16.5 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 1.8 |
7.1 |
GO:1900623 |
regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 1.8 |
5.3 |
GO:1904685 |
positive regulation of metalloendopeptidase activity(GO:1904685) |
| 1.7 |
8.5 |
GO:0035426 |
extracellular matrix-cell signaling(GO:0035426) |
| 1.4 |
4.3 |
GO:0086046 |
membrane depolarization during SA node cell action potential(GO:0086046) |
| 1.4 |
8.4 |
GO:2000672 |
negative regulation of motor neuron apoptotic process(GO:2000672) |
| 1.4 |
5.5 |
GO:0034975 |
protein folding in endoplasmic reticulum(GO:0034975) |
| 1.2 |
8.5 |
GO:0032811 |
negative regulation of gamma-aminobutyric acid secretion(GO:0014053) negative regulation of epinephrine secretion(GO:0032811) |
| 1.2 |
5.9 |
GO:0051045 |
negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 1.0 |
3.1 |
GO:0006558 |
L-phenylalanine metabolic process(GO:0006558) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) |
| 1.0 |
3.0 |
GO:0016191 |
synaptic vesicle uncoating(GO:0016191) regulation of endosome organization(GO:1904978) positive regulation of endosome organization(GO:1904980) |
| 1.0 |
4.8 |
GO:0050862 |
positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.9 |
2.8 |
GO:1902460 |
regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.9 |
6.3 |
GO:0007342 |
fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.9 |
9.8 |
GO:0071635 |
negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.9 |
2.6 |
GO:0042138 |
meiotic DNA double-strand break formation(GO:0042138) |
| 0.8 |
3.4 |
GO:2000823 |
regulation of androgen receptor activity(GO:2000823) |
| 0.8 |
5.9 |
GO:0030579 |
ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.8 |
2.4 |
GO:0043696 |
dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.8 |
3.9 |
GO:0001661 |
conditioned taste aversion(GO:0001661) |
| 0.8 |
8.3 |
GO:0046543 |
development of secondary female sexual characteristics(GO:0046543) |
| 0.8 |
0.8 |
GO:0003221 |
right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.8 |
9.0 |
GO:1902993 |
positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.7 |
2.9 |
GO:0010808 |
positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.7 |
7.2 |
GO:2001214 |
positive regulation of vasculogenesis(GO:2001214) |
| 0.7 |
2.1 |
GO:0019676 |
ammonia assimilation cycle(GO:0019676) |
| 0.7 |
4.8 |
GO:0021940 |
positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.7 |
4.1 |
GO:1904936 |
cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.7 |
8.0 |
GO:2000465 |
regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.7 |
6.6 |
GO:0045741 |
positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.6 |
7.7 |
GO:0042711 |
maternal behavior(GO:0042711) |
| 0.6 |
1.9 |
GO:0006533 |
aspartate catabolic process(GO:0006533) D-amino acid catabolic process(GO:0019478) |
| 0.6 |
2.5 |
GO:0086045 |
smooth muscle contraction involved in micturition(GO:0060083) membrane depolarization during AV node cell action potential(GO:0086045) membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.6 |
2.4 |
GO:1903587 |
regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.6 |
2.4 |
GO:0097167 |
circadian regulation of translation(GO:0097167) |
| 0.6 |
11.4 |
GO:0021516 |
dorsal spinal cord development(GO:0021516) |
| 0.6 |
3.5 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.6 |
2.3 |
GO:0007262 |
STAT protein import into nucleus(GO:0007262) |
| 0.6 |
1.7 |
GO:0021914 |
negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.6 |
0.6 |
GO:0060681 |
branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.6 |
3.5 |
GO:0034393 |
positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.6 |
3.4 |
GO:0009449 |
gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.5 |
4.9 |
GO:0036465 |
synaptic vesicle recycling(GO:0036465) |
| 0.5 |
3.2 |
GO:0070863 |
positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.5 |
2.7 |
GO:1904721 |
regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.5 |
1.6 |
GO:0045204 |
MAPK export from nucleus(GO:0045204) |
| 0.5 |
3.0 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.5 |
2.5 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 0.5 |
1.9 |
GO:0061343 |
cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.5 |
1.4 |
GO:0070589 |
cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule metabolic process(GO:0044036) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) cell wall organization or biogenesis(GO:0071554) |
| 0.5 |
1.9 |
GO:0009957 |
epidermal cell fate specification(GO:0009957) |
| 0.5 |
2.8 |
GO:0071372 |
cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.5 |
2.3 |
GO:0032534 |
regulation of microvillus assembly(GO:0032534) |
| 0.5 |
2.3 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.4 |
3.6 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.4 |
4.0 |
GO:0071435 |
potassium ion export(GO:0071435) |
| 0.4 |
1.6 |
GO:1904425 |
negative regulation of GTP binding(GO:1904425) |
| 0.4 |
6.6 |
GO:0001553 |
luteinization(GO:0001553) |
| 0.4 |
18.6 |
GO:0007528 |
neuromuscular junction development(GO:0007528) |
| 0.4 |
4.4 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 0.4 |
14.6 |
GO:0000038 |
very long-chain fatty acid metabolic process(GO:0000038) |
| 0.4 |
3.0 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.4 |
1.4 |
GO:2000812 |
regulation of barbed-end actin filament capping(GO:2000812) |
| 0.3 |
1.0 |
GO:1903722 |
regulation of centriole elongation(GO:1903722) |
| 0.3 |
1.7 |
GO:0010961 |
cellular magnesium ion homeostasis(GO:0010961) |
| 0.3 |
2.3 |
GO:2000807 |
regulation of synaptic vesicle clustering(GO:2000807) |
| 0.3 |
1.6 |
GO:0030259 |
lipid glycosylation(GO:0030259) |
| 0.3 |
5.1 |
GO:0031958 |
corticosteroid receptor signaling pathway(GO:0031958) |
| 0.3 |
0.3 |
GO:0048023 |
positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.3 |
0.9 |
GO:1902358 |
sulfate transmembrane transport(GO:1902358) |
| 0.3 |
2.1 |
GO:0015840 |
urea transport(GO:0015840) urea transmembrane transport(GO:0071918) |
| 0.3 |
0.9 |
GO:1904742 |
protein poly-ADP-ribosylation(GO:0070212) regulation of telomeric DNA binding(GO:1904742) |
| 0.3 |
20.2 |
GO:0048663 |
neuron fate commitment(GO:0048663) |
| 0.3 |
1.6 |
GO:0060872 |
semicircular canal morphogenesis(GO:0048752) semicircular canal development(GO:0060872) |
| 0.3 |
2.8 |
GO:0045723 |
positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.3 |
1.8 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.3 |
2.3 |
GO:0015868 |
purine ribonucleotide transport(GO:0015868) |
| 0.2 |
1.7 |
GO:0000050 |
urea cycle(GO:0000050) urea metabolic process(GO:0019627) cellular response to lithium ion(GO:0071285) |
| 0.2 |
0.5 |
GO:0010512 |
negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.2 |
1.0 |
GO:0055071 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.2 |
1.0 |
GO:0019276 |
UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.2 |
1.4 |
GO:0021636 |
trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) |
| 0.2 |
6.5 |
GO:0051491 |
positive regulation of filopodium assembly(GO:0051491) |
| 0.2 |
8.4 |
GO:2001258 |
negative regulation of cation channel activity(GO:2001258) |
| 0.2 |
2.0 |
GO:0097119 |
postsynaptic density protein 95 clustering(GO:0097119) |
| 0.2 |
1.4 |
GO:0036233 |
glycine import(GO:0036233) |
| 0.2 |
1.1 |
GO:0061687 |
detoxification of inorganic compound(GO:0061687) |
| 0.2 |
0.8 |
GO:0046487 |
glyoxylate metabolic process(GO:0046487) |
| 0.2 |
5.4 |
GO:2000463 |
positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.2 |
1.3 |
GO:0045200 |
establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.2 |
2.2 |
GO:2000766 |
negative regulation of cytoplasmic translation(GO:2000766) |
| 0.2 |
0.4 |
GO:0003032 |
detection of oxygen(GO:0003032) |
| 0.2 |
1.4 |
GO:0019348 |
dolichol metabolic process(GO:0019348) |
| 0.2 |
0.7 |
GO:0072365 |
regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.2 |
1.2 |
GO:0099625 |
regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.2 |
1.0 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 |
0.3 |
GO:2001025 |
positive regulation of response to drug(GO:2001025) |
| 0.2 |
0.6 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.1 |
1.3 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.1 |
0.4 |
GO:0021523 |
somatic motor neuron differentiation(GO:0021523) |
| 0.1 |
1.0 |
GO:0048681 |
negative regulation of axon regeneration(GO:0048681) |
| 0.1 |
0.9 |
GO:1901844 |
regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.1 |
9.3 |
GO:0072583 |
clathrin-mediated endocytosis(GO:0072583) |
| 0.1 |
0.7 |
GO:0048597 |
post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.1 |
0.7 |
GO:0090164 |
asymmetric Golgi ribbon formation(GO:0090164) |
| 0.1 |
1.4 |
GO:0051601 |
exocyst localization(GO:0051601) |
| 0.1 |
0.8 |
GO:0001542 |
ovulation from ovarian follicle(GO:0001542) |
| 0.1 |
0.4 |
GO:1900275 |
negative regulation of phospholipase C activity(GO:1900275) regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 0.1 |
5.4 |
GO:0032418 |
lysosome localization(GO:0032418) |
| 0.1 |
2.3 |
GO:0035094 |
response to nicotine(GO:0035094) |
| 0.1 |
2.8 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 |
0.3 |
GO:0070375 |
ERK5 cascade(GO:0070375) |
| 0.1 |
0.6 |
GO:0051697 |
protein delipidation(GO:0051697) |
| 0.1 |
0.4 |
GO:0019042 |
viral latency(GO:0019042) |
| 0.1 |
1.7 |
GO:0007216 |
G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 |
0.6 |
GO:0043615 |
astrocyte cell migration(GO:0043615) |
| 0.1 |
0.8 |
GO:0061088 |
sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 |
3.7 |
GO:0007520 |
myoblast fusion(GO:0007520) |
| 0.1 |
0.7 |
GO:0006498 |
N-terminal protein lipidation(GO:0006498) |
| 0.1 |
1.3 |
GO:0048305 |
immunoglobulin secretion(GO:0048305) |
| 0.1 |
3.1 |
GO:0090004 |
positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 |
5.2 |
GO:0030216 |
keratinocyte differentiation(GO:0030216) |
| 0.1 |
0.2 |
GO:0010726 |
positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.1 |
0.7 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 |
6.4 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.1 |
0.9 |
GO:0097011 |
cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.1 |
1.4 |
GO:0070207 |
protein homotrimerization(GO:0070207) |
| 0.1 |
0.3 |
GO:0035864 |
response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) axonogenesis involved in innervation(GO:0060385) |
| 0.1 |
2.0 |
GO:0033198 |
response to ATP(GO:0033198) |
| 0.1 |
1.5 |
GO:0090083 |
regulation of inclusion body assembly(GO:0090083) |
| 0.1 |
1.4 |
GO:0051898 |
negative regulation of protein kinase B signaling(GO:0051898) |
| 0.1 |
1.3 |
GO:0035024 |
negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 |
5.2 |
GO:0043407 |
negative regulation of MAP kinase activity(GO:0043407) |
| 0.1 |
3.5 |
GO:0031529 |
ruffle organization(GO:0031529) |
| 0.1 |
0.2 |
GO:0003256 |
regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003256) atrioventricular canal development(GO:0036302) |
| 0.1 |
0.9 |
GO:2001197 |
regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 |
1.5 |
GO:0032981 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 |
1.6 |
GO:0046329 |
negative regulation of JNK cascade(GO:0046329) |
| 0.1 |
0.9 |
GO:0060384 |
innervation(GO:0060384) |
| 0.1 |
1.2 |
GO:2000311 |
regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 |
0.2 |
GO:0045950 |
telomeric loop formation(GO:0031627) negative regulation of mitotic recombination(GO:0045950) negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.1 |
0.8 |
GO:0014067 |
negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.1 |
2.3 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.1 |
1.4 |
GO:0070266 |
necroptotic process(GO:0070266) |
| 0.1 |
0.4 |
GO:0050882 |
voluntary musculoskeletal movement(GO:0050882) |
| 0.1 |
1.8 |
GO:0035418 |
protein localization to synapse(GO:0035418) |
| 0.1 |
0.2 |
GO:0070124 |
mitochondrial translational initiation(GO:0070124) |
| 0.1 |
2.9 |
GO:0045839 |
negative regulation of mitotic nuclear division(GO:0045839) |
| 0.1 |
3.9 |
GO:0051965 |
positive regulation of synapse assembly(GO:0051965) |
| 0.1 |
2.5 |
GO:0006893 |
Golgi to plasma membrane transport(GO:0006893) |
| 0.1 |
0.7 |
GO:0007095 |
mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 |
0.4 |
GO:0044539 |
long-chain fatty acid import(GO:0044539) |
| 0.1 |
2.0 |
GO:0021799 |
cerebral cortex radially oriented cell migration(GO:0021799) |
| 0.1 |
0.3 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.1 |
2.6 |
GO:0061077 |
chaperone-mediated protein folding(GO:0061077) |
| 0.1 |
7.2 |
GO:0006813 |
potassium ion transport(GO:0006813) |
| 0.1 |
0.3 |
GO:0071557 |
histone H3-K27 demethylation(GO:0071557) |
| 0.0 |
1.8 |
GO:0072661 |
protein targeting to plasma membrane(GO:0072661) |
| 0.0 |
2.1 |
GO:0050729 |
positive regulation of inflammatory response(GO:0050729) |
| 0.0 |
0.5 |
GO:0033169 |
histone H3-K9 demethylation(GO:0033169) regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 |
0.4 |
GO:0070914 |
UV-damage excision repair(GO:0070914) |
| 0.0 |
1.0 |
GO:0045494 |
photoreceptor cell maintenance(GO:0045494) |
| 0.0 |
0.5 |
GO:0007289 |
spermatid nucleus differentiation(GO:0007289) |
| 0.0 |
1.1 |
GO:2000785 |
regulation of autophagosome assembly(GO:2000785) |
| 0.0 |
0.2 |
GO:0035428 |
hexose transmembrane transport(GO:0035428) |
| 0.0 |
0.4 |
GO:0006828 |
manganese ion transport(GO:0006828) |
| 0.0 |
0.2 |
GO:0010919 |
regulation of inositol phosphate biosynthetic process(GO:0010919) positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 |
0.3 |
GO:0042407 |
cristae formation(GO:0042407) |
| 0.0 |
1.2 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
0.1 |
GO:0032926 |
negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 |
0.1 |
GO:0006703 |
estrogen biosynthetic process(GO:0006703) |
| 0.0 |
0.1 |
GO:0006361 |
transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 |
0.5 |
GO:0031146 |
SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 |
1.1 |
GO:0035383 |
acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 |
0.1 |
GO:0071985 |
multivesicular body sorting pathway(GO:0071985) |
| 0.0 |
0.3 |
GO:0045670 |
regulation of osteoclast differentiation(GO:0045670) |
| 0.0 |
0.6 |
GO:0048286 |
lung alveolus development(GO:0048286) |
| 0.0 |
0.4 |
GO:0006506 |
GPI anchor biosynthetic process(GO:0006506) |