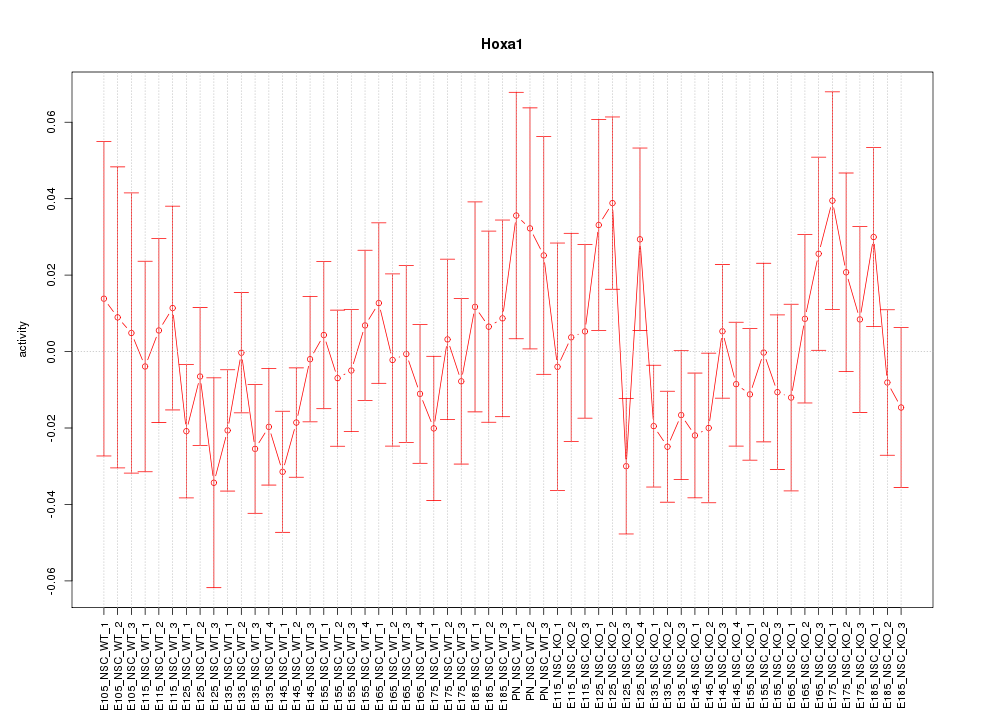
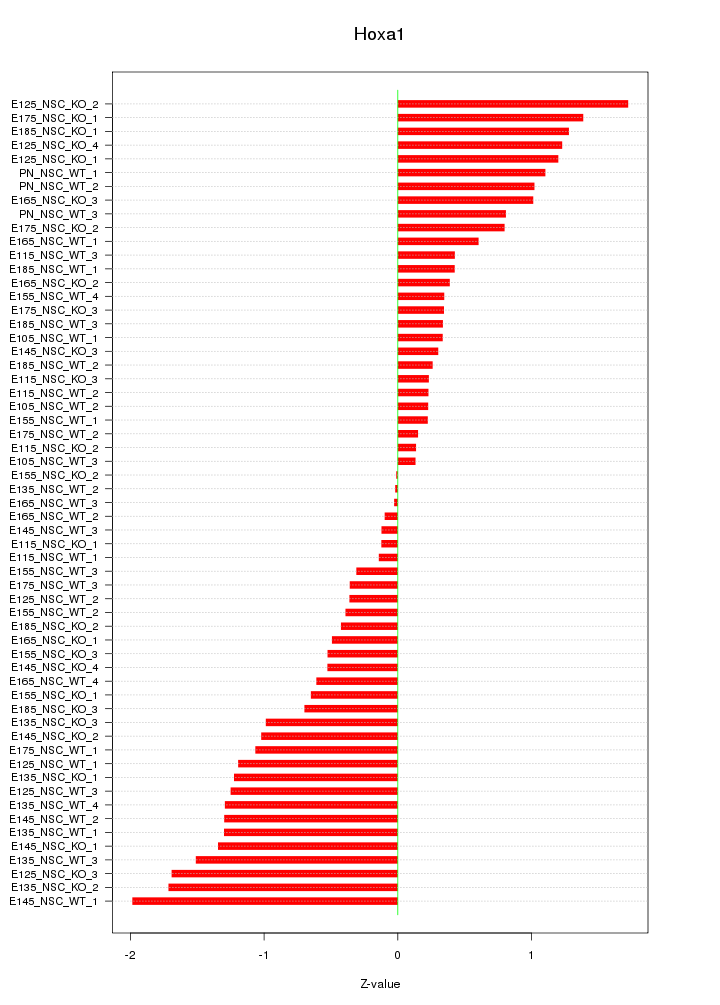
Motif ID: Hoxa1
Z-value: 0.875
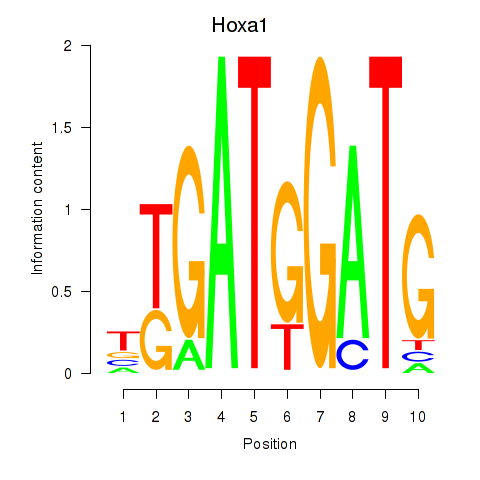
Transcription factors associated with Hoxa1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hoxa1 | ENSMUSG00000029844.9 | Hoxa1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.7 | 2.0 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.7 | 2.0 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.6 | 1.9 | GO:0051695 | actin filament uncapping(GO:0051695) |
| 0.6 | 1.8 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.6 | 1.8 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.5 | 3.4 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.4 | 4.8 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.4 | 4.8 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.4 | 1.1 | GO:0045014 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.3 | 3.9 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.3 | 0.9 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.3 | 1.4 | GO:0046864 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) alveolar primary septum development(GO:0061143) |
| 0.2 | 2.2 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.2 | 0.7 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.2 | 3.9 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.2 | 1.5 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.2 | 1.8 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.1 | 1.3 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.1 | 0.5 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 1.1 | GO:0043084 | penile erection(GO:0043084) |
| 0.1 | 1.2 | GO:0001553 | luteinization(GO:0001553) |
| 0.1 | 0.4 | GO:0035989 | tendon development(GO:0035989) |
| 0.1 | 0.5 | GO:0042711 | maternal behavior(GO:0042711) |
| 0.0 | 1.3 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 2.1 | GO:1901381 | positive regulation of potassium ion transmembrane transport(GO:1901381) |
| 0.0 | 2.1 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.6 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.5 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.8 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 1.3 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 2.7 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 0.6 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.0 | 0.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.3 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.7 | GO:0006278 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.0 | 2.0 | GO:0002757 | immune response-activating signal transduction(GO:0002757) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.4 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.6 | 1.8 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 1.3 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.2 | 0.6 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 1.9 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 0.8 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.4 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 3.7 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 2.9 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.0 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 1.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.1 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 4.0 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 1.8 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.4 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.9 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 2.8 | GO:0031965 | nuclear membrane(GO:0031965) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.4 | 2.5 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.4 | 4.2 | GO:0008517 | folic acid transporter activity(GO:0008517) water channel activity(GO:0015250) |
| 0.2 | 1.9 | GO:0031432 | thyroid hormone receptor coactivator activity(GO:0030375) titin binding(GO:0031432) |
| 0.2 | 3.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 0.9 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.2 | 2.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 1.5 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) L-proline transmembrane transporter activity(GO:0015193) alanine transmembrane transporter activity(GO:0022858) |
| 0.2 | 1.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.2 | 0.5 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 0.2 | 2.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.2 | 0.7 | GO:0004096 | catalase activity(GO:0004096) |
| 0.2 | 2.0 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 3.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 2.4 | GO:0034946 | 2-oxoglutaryl-CoA thioesterase activity(GO:0034843) 2,4,4-trimethyl-3-oxopentanoyl-CoA thioesterase activity(GO:0034869) 3-isopropylbut-3-enoyl-CoA thioesterase activity(GO:0034946) glutaryl-CoA hydrolase activity(GO:0044466) |
| 0.1 | 1.3 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 0.5 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.1 | 1.4 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.1 | 2.0 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 1.8 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 0.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 0.6 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 1.1 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 2.1 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 4.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 1.1 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 1.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.7 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 3.5 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.8 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |