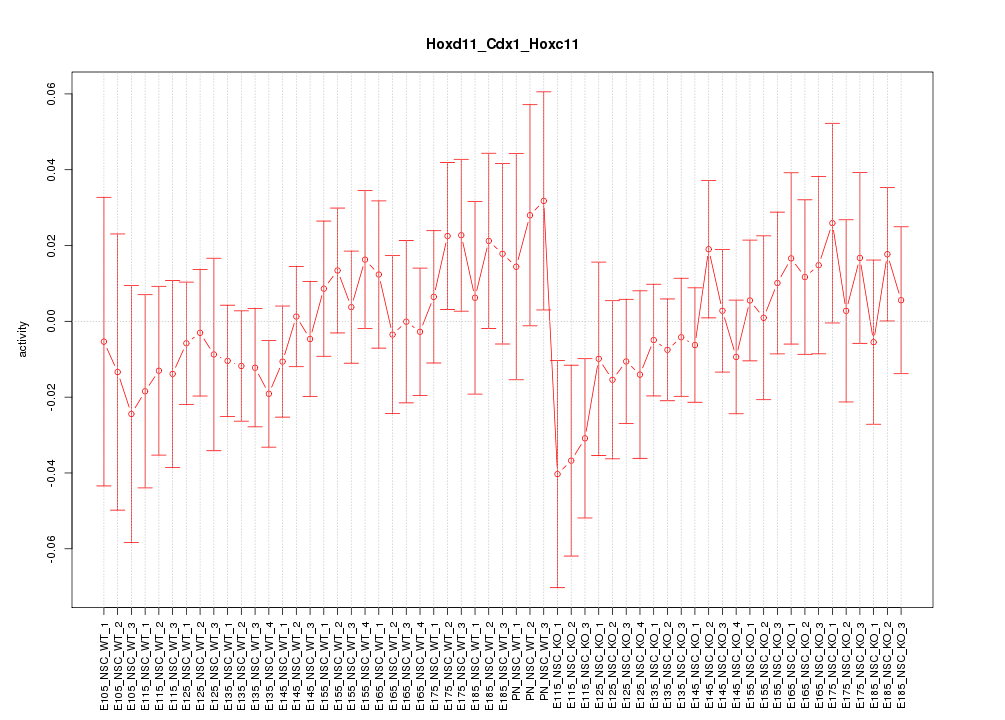
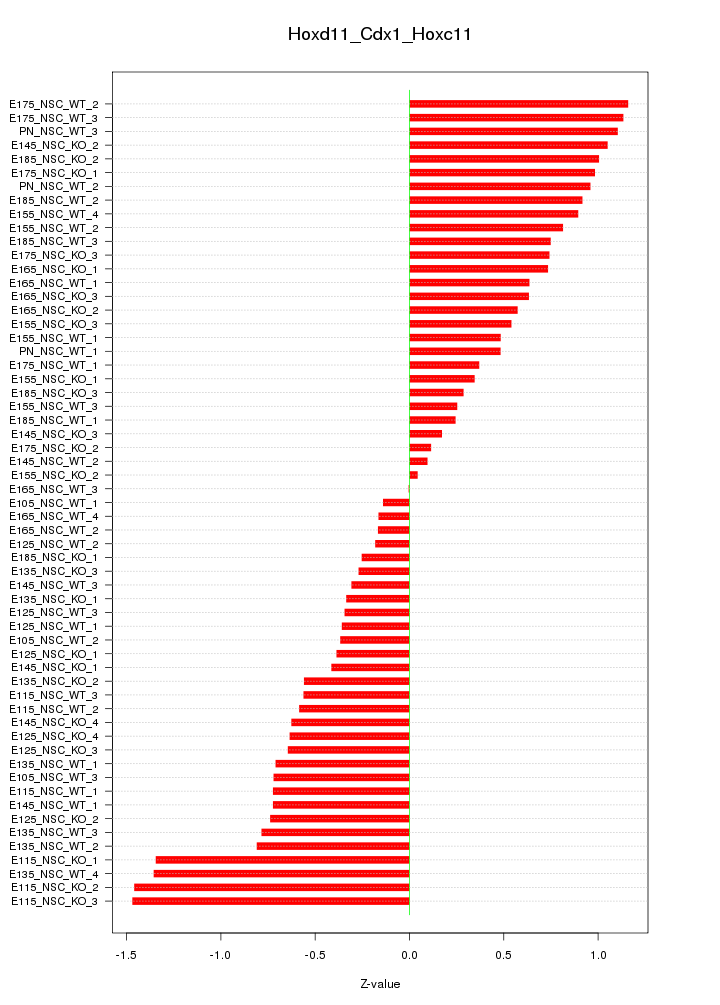
Motif ID: Hoxd11_Cdx1_Hoxc11
Z-value: 0.706
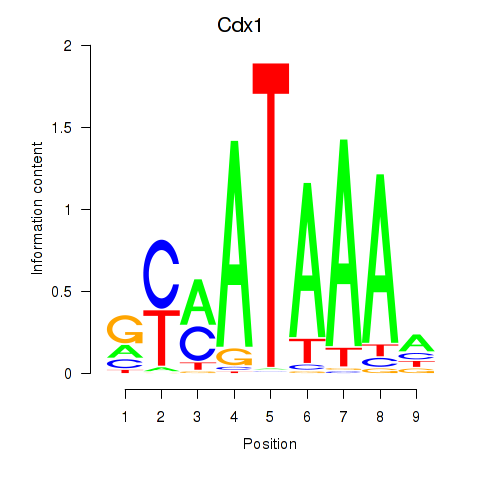
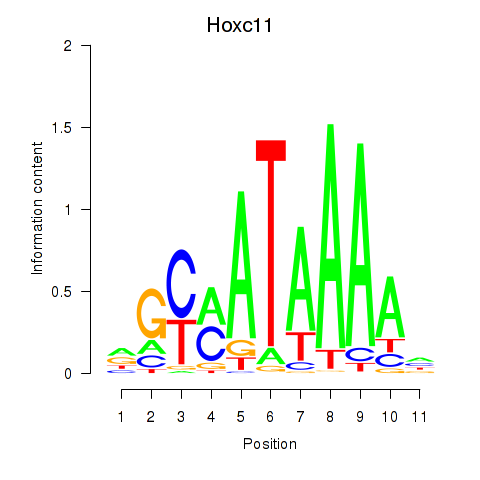
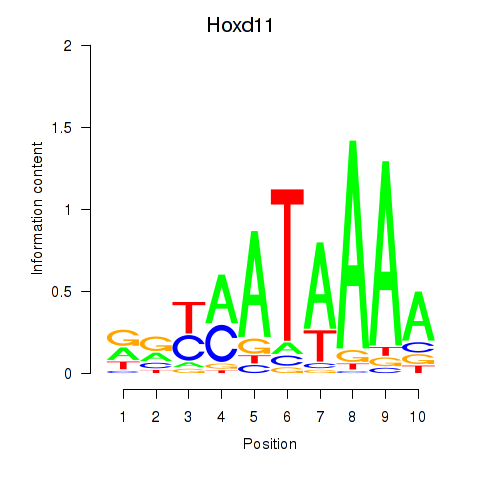
Transcription factors associated with Hoxd11_Cdx1_Hoxc11:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Cdx1 | ENSMUSG00000024619.8 | Cdx1 |
| Hoxc11 | ENSMUSG00000001656.3 | Hoxc11 |
| Hoxd11 | ENSMUSG00000042499.12 | Hoxd11 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.8 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 2.1 | 10.6 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 1.9 | 21.1 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 1.3 | 5.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.7 | 4.7 | GO:0034047 | regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.6 | 2.4 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.6 | 2.9 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.5 | 2.5 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.4 | 4.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.4 | 4.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.4 | 2.0 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.4 | 2.7 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.4 | 1.1 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.3 | 1.0 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.3 | 2.0 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.3 | 0.9 | GO:0061043 | regulation of vascular wound healing(GO:0061043) |
| 0.3 | 0.9 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.3 | 1.2 | GO:2001137 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) positive regulation of endocytic recycling(GO:2001137) |
| 0.3 | 1.1 | GO:0060745 | mammary gland branching involved in pregnancy(GO:0060745) |
| 0.3 | 4.3 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.3 | 3.1 | GO:1901629 | regulation of presynaptic membrane organization(GO:1901629) |
| 0.2 | 0.4 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.2 | 1.1 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.2 | 1.1 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.2 | 1.3 | GO:1903056 | regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.2 | 2.0 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 0.8 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.2 | 0.6 | GO:0008228 | opsonization(GO:0008228) modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.2 | 1.5 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.2 | 4.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.2 | 0.6 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.2 | 0.5 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.2 | 0.5 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.2 | 1.8 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 2.7 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.6 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 0.4 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 2.5 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 1.2 | GO:0007379 | segment specification(GO:0007379) |
| 0.1 | 2.1 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.1 | 0.5 | GO:0032346 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.1 | 0.2 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.1 | 0.4 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 2.8 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 0.8 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.1 | 0.9 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 | 0.6 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 | 0.3 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 7.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 1.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.6 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.8 | GO:1900153 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.4 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.4 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.8 | GO:0071398 | cellular response to fatty acid(GO:0071398) |
| 0.0 | 0.9 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.1 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.7 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 3.9 | GO:0045814 | negative regulation of gene expression, epigenetic(GO:0045814) |
| 0.0 | 0.3 | GO:0036120 | response to platelet-derived growth factor(GO:0036119) cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 | 0.6 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.7 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.0 | 15.1 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.0 | 0.2 | GO:0002674 | negative regulation of acute inflammatory response(GO:0002674) |
| 0.0 | 0.4 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.3 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.3 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.0 | GO:0009838 | abscission(GO:0009838) |
| 0.0 | 0.3 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.3 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.0 | 0.3 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.9 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.0 | 0.6 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.2 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.4 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 1.3 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.3 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.8 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.1 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.2 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.8 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.6 | 2.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.3 | 5.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 5.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 5.1 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 10.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 0.4 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 0.5 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 1.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 4.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 7.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 3.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 1.1 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 5.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 2.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.3 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.9 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 1.2 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 2.4 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 1.1 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 2.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.0 | 0.4 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.4 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 1.5 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 1.7 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 0.6 | GO:0072686 | mitotic spindle(GO:0072686) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 21.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 1.1 | 4.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 1.0 | 4.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.9 | 10.6 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.8 | 5.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.8 | 11.8 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.6 | 2.5 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.6 | 2.4 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.6 | 2.9 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.4 | 1.1 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.3 | 4.7 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.3 | 1.0 | GO:0008061 | chitin binding(GO:0008061) |
| 0.2 | 12.0 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.2 | 0.5 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.4 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.1 | 0.9 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 2.7 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 3.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 2.2 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 1.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 0.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 1.5 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.1 | 1.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.3 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 12.8 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.1 | 0.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 1.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.3 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 0.5 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.9 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.0 | 1.1 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.7 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 1.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.6 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 3.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 1.5 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 1.2 | GO:0034945 | dihydrolipoamide branched chain acyltransferase activity(GO:0004147) palmitoleoyl [acyl-carrier-protein]-dependent acyltransferase activity(GO:0008951) serine O-acyltransferase activity(GO:0016412) O-succinyltransferase activity(GO:0016750) sinapoyltransferase activity(GO:0016752) O-sinapoyltransferase activity(GO:0016753) peptidyl-lysine N6-myristoyltransferase activity(GO:0018030) peptidyl-lysine N6-palmitoyltransferase activity(GO:0018031) benzoyl acetate-CoA thiolase activity(GO:0018711) 3-hydroxybutyryl-CoA thiolase activity(GO:0018712) 3-ketopimelyl-CoA thiolase activity(GO:0018713) N-palmitoyltransferase activity(GO:0019105) acyl-CoA N-acyltransferase activity(GO:0019186) protein-cysteine S-myristoyltransferase activity(GO:0019705) glucosaminyl-phosphotidylinositol O-acyltransferase activity(GO:0032216) ergosterol O-acyltransferase activity(GO:0034737) lanosterol O-acyltransferase activity(GO:0034738) naphthyl-2-oxomethyl-succinyl-CoA succinyl transferase activity(GO:0034848) 2,4,4-trimethyl-3-oxopentanoyl-CoA 2-C-propanoyl transferase activity(GO:0034851) 2-methylhexanoyl-CoA C-acetyltransferase activity(GO:0034915) butyryl-CoA 2-C-propionyltransferase activity(GO:0034919) 2,6-dimethyl-5-methylene-3-oxo-heptanoyl-CoA C-acetyltransferase activity(GO:0034945) L-2-aminoadipate N-acetyltransferase activity(GO:0043741) keto acid formate lyase activity(GO:0043806) azetidine-2-carboxylic acid acetyltransferase activity(GO:0046941) peptidyl-lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0052858) acetyl-CoA:L-lysine N6-acetyltransferase(GO:0090595) |
| 0.0 | 0.7 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 1.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.4 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.4 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.4 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.1 | GO:0034511 | U4 snRNA binding(GO:0030621) U3 snoRNA binding(GO:0034511) |
| 0.0 | 10.7 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 0.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.5 | GO:0061733 | peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 1.3 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.5 | GO:0031490 | chromatin DNA binding(GO:0031490) |