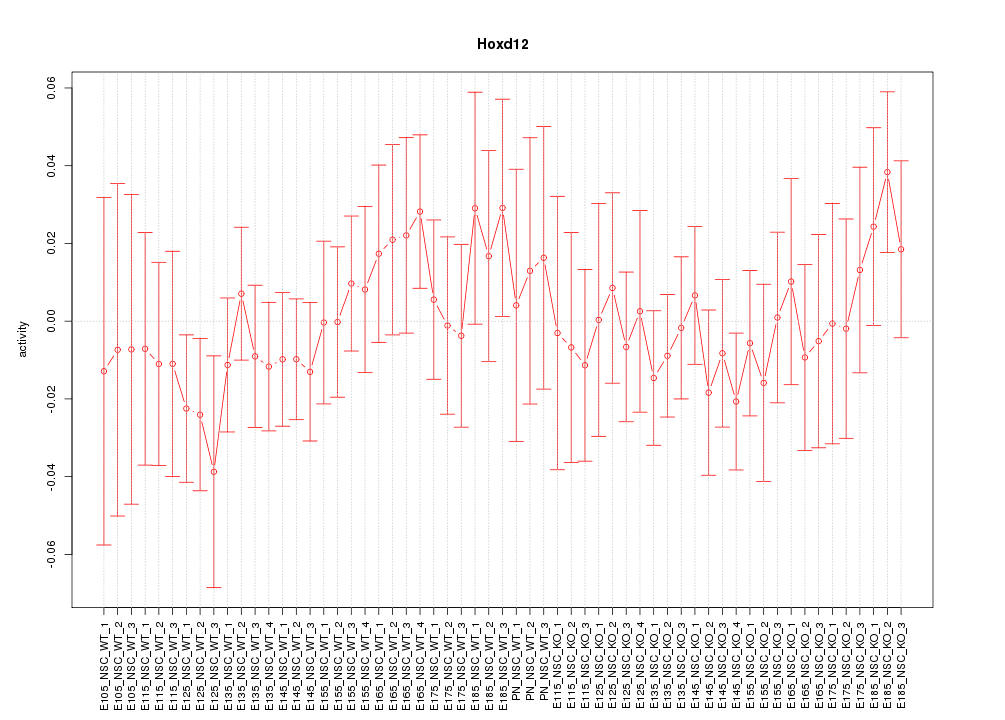
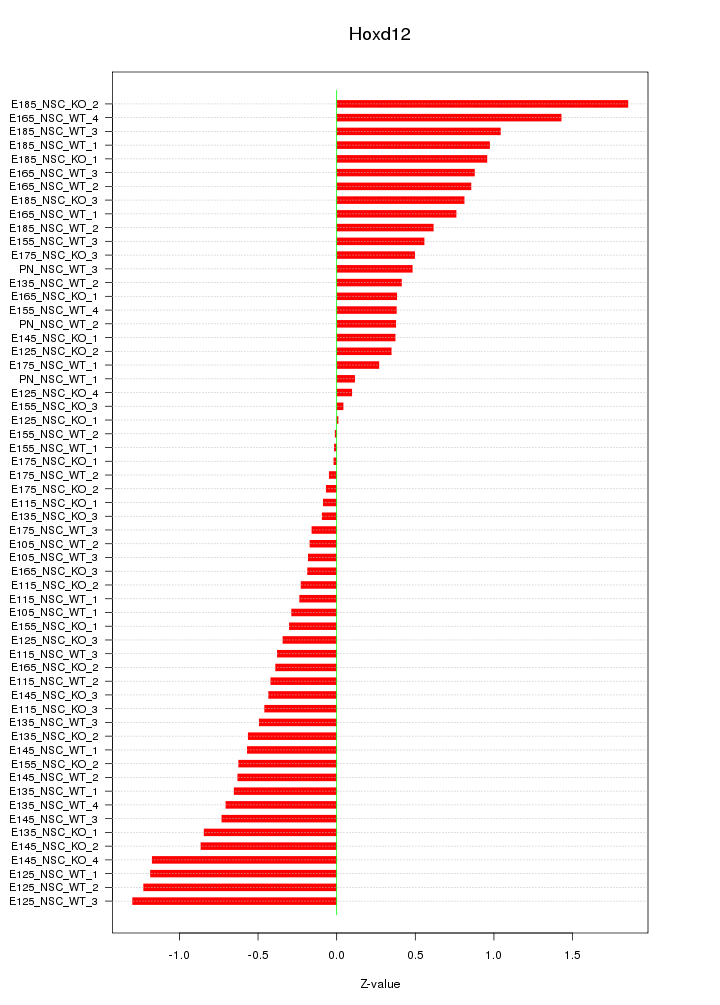
Motif ID: Hoxd12
Z-value: 0.655

Transcription factors associated with Hoxd12:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hoxd12 | ENSMUSG00000001823.4 | Hoxd12 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 11.7 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 1.8 | 5.5 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 1.1 | 12.0 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.5 | 2.7 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.5 | 1.9 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.4 | 2.0 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.3 | 5.2 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.2 | 2.5 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.2 | 0.7 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.2 | 0.9 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.2 | 1.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.2 | 1.0 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.2 | 1.6 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.2 | 1.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.2 | 0.5 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.2 | 3.0 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.6 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 4.2 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 2.2 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 1.0 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 3.9 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.5 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.1 | 0.3 | GO:0061043 | regulation of vascular wound healing(GO:0061043) |
| 0.1 | 0.6 | GO:1903056 | regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.1 | 2.3 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.1 | 0.9 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 1.7 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.1 | 0.8 | GO:0007379 | segment specification(GO:0007379) |
| 0.1 | 2.1 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 0.6 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 1.1 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.4 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.5 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 2.7 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.3 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.5 | GO:1900153 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.6 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.2 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.2 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.0 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 5.5 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.0 | 0.9 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.3 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.7 | 11.7 | GO:0043205 | fibril(GO:0043205) |
| 0.2 | 0.9 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.6 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 2.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 0.5 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.1 | 0.5 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 1.9 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 1.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.6 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 6.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 2.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 1.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 2.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.5 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.8 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 1.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 3.8 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 1.1 | GO:0016605 | PML body(GO:0016605) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 11.7 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 1.7 | 12.0 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.6 | 2.5 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.4 | 2.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.4 | 5.5 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.2 | 0.9 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.2 | 2.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 2.7 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.2 | 1.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.2 | 3.0 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.2 | 7.6 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.6 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 2.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 1.7 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 0.6 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 1.0 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 1.0 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.0 | 0.7 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.0 | 0.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.6 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 1.1 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.5 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 2.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.9 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 4.0 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.3 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 0.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.7 | GO:0046332 | SMAD binding(GO:0046332) |