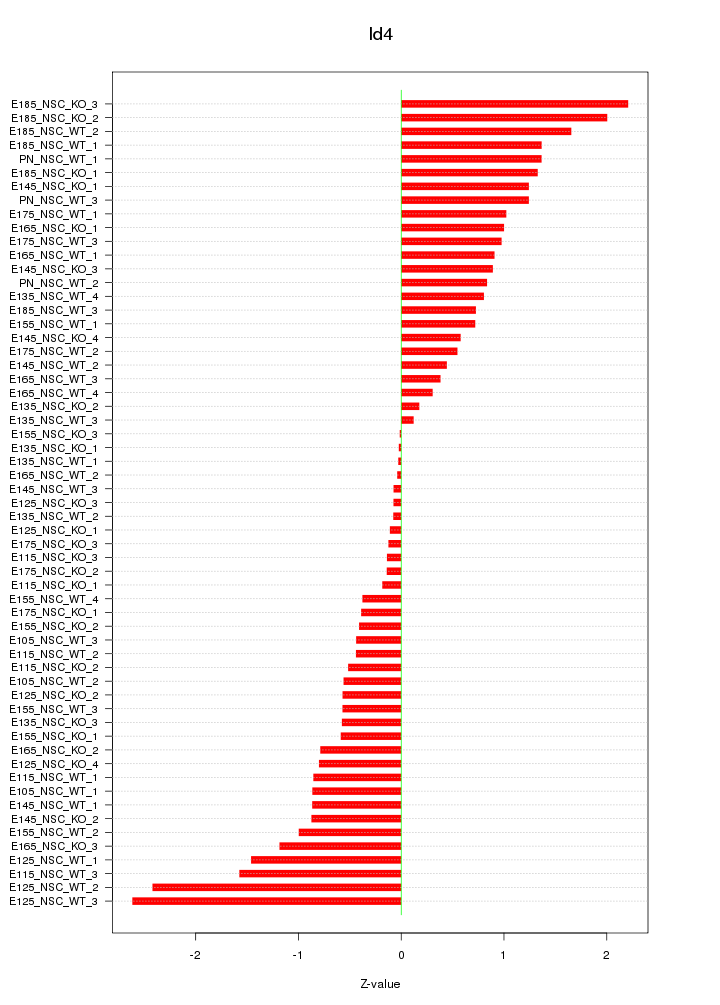
Motif ID: Id4
Z-value: 0.967
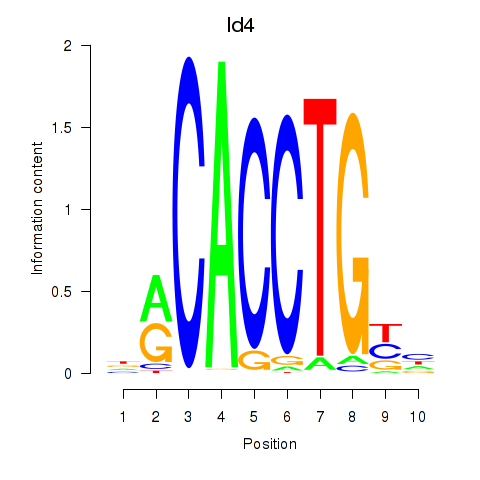
Transcription factors associated with Id4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Id4 | ENSMUSG00000021379.1 | Id4 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Id4 | mm10_v2_chr13_+_48261427_48261427 | -0.13 | 3.1e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.5 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 3.4 | 13.6 | GO:1903288 | regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 2.8 | 8.4 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 2.4 | 7.2 | GO:1904049 | regulation of spontaneous neurotransmitter secretion(GO:1904048) negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 2.2 | 8.7 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 2.0 | 5.9 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 1.7 | 1.7 | GO:0090194 | negative regulation of glomerular mesangial cell proliferation(GO:0072125) negative regulation of glomerulus development(GO:0090194) |
| 1.6 | 8.1 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) |
| 1.6 | 6.3 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 1.6 | 4.7 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 1.5 | 4.6 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 1.4 | 4.3 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 1.4 | 2.7 | GO:1903275 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) |
| 1.3 | 6.6 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 1.2 | 4.7 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 1.1 | 3.4 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 1.1 | 3.4 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 1.1 | 5.7 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 1.1 | 6.6 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 1.0 | 4.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 1.0 | 1.0 | GO:1990379 | lipid transport across blood brain barrier(GO:1990379) |
| 1.0 | 3.0 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 1.0 | 6.9 | GO:0034047 | regulation of protein phosphatase type 2A activity(GO:0034047) |
| 1.0 | 1.0 | GO:2000823 | regulation of androgen receptor activity(GO:2000823) |
| 0.9 | 2.8 | GO:0046959 | habituation(GO:0046959) |
| 0.9 | 3.6 | GO:0033580 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.9 | 3.6 | GO:0060032 | notochord regression(GO:0060032) |
| 0.9 | 2.7 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.8 | 2.4 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 0.8 | 3.9 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.8 | 5.4 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.8 | 2.3 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.8 | 3.1 | GO:0010046 | arginine biosynthetic process(GO:0006526) response to mycotoxin(GO:0010046) |
| 0.7 | 4.3 | GO:0045188 | optic nerve morphogenesis(GO:0021631) regulation of circadian sleep/wake cycle, non-REM sleep(GO:0045188) |
| 0.7 | 4.2 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.7 | 2.0 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.7 | 2.0 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.6 | 3.1 | GO:0075136 | response to defenses of other organism involved in symbiotic interaction(GO:0052173) response to host defenses(GO:0052200) response to host(GO:0075136) |
| 0.6 | 2.4 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.6 | 2.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.6 | 1.7 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.6 | 1.7 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.6 | 3.3 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.5 | 2.7 | GO:0015871 | choline transport(GO:0015871) |
| 0.5 | 1.1 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.5 | 13.3 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.5 | 3.7 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.5 | 3.6 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.5 | 7.3 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.5 | 2.0 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.5 | 1.4 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.5 | 1.8 | GO:1902512 | positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.5 | 2.8 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.5 | 4.1 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.4 | 3.1 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.4 | 1.7 | GO:1903181 | regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 0.4 | 5.9 | GO:0042182 | ketone catabolic process(GO:0042182) |
| 0.4 | 1.2 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.4 | 1.6 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.4 | 3.6 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.4 | 1.6 | GO:1902965 | protein localization to early endosome(GO:1902946) positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.4 | 1.2 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.4 | 1.2 | GO:1904395 | positive regulation of receptor clustering(GO:1903911) regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.4 | 3.9 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.4 | 0.8 | GO:0051464 | positive regulation of cortisol secretion(GO:0051464) |
| 0.4 | 1.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.4 | 1.5 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.4 | 3.4 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.4 | 4.1 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.4 | 3.3 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.4 | 2.8 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.3 | 0.7 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.3 | 1.0 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.3 | 2.0 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.3 | 1.4 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) closure of optic fissure(GO:0061386) |
| 0.3 | 1.3 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.3 | 1.7 | GO:0071476 | cellular hypotonic response(GO:0071476) |
| 0.3 | 4.3 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.3 | 2.0 | GO:0002591 | positive regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002591) |
| 0.3 | 3.8 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.3 | 3.8 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.3 | 1.5 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.3 | 0.6 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.3 | 3.0 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.3 | 3.6 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.3 | 3.6 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.3 | 1.5 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.3 | 4.8 | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) |
| 0.3 | 4.1 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.3 | 1.7 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.3 | 1.4 | GO:0032346 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.3 | 2.5 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.3 | 3.9 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.3 | 2.8 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.3 | 0.5 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.3 | 1.6 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.3 | 1.0 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.3 | 6.3 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.3 | 3.4 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.3 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.3 | 4.6 | GO:0008209 | androgen metabolic process(GO:0008209) |
| 0.3 | 1.3 | GO:0090032 | negative regulation of steroid hormone biosynthetic process(GO:0090032) |
| 0.3 | 2.3 | GO:0030432 | peristalsis(GO:0030432) |
| 0.3 | 0.8 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.3 | 1.5 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.2 | 1.5 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.2 | 1.7 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.2 | 0.7 | GO:1903061 | glutamate secretion, neurotransmission(GO:0061535) establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of protein lipidation(GO:1903059) positive regulation of protein lipidation(GO:1903061) |
| 0.2 | 2.9 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.2 | 1.9 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.2 | 1.0 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.2 | 3.1 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 1.7 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.2 | 2.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 3.0 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.2 | 1.6 | GO:0045187 | regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.2 | 4.8 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.2 | 1.4 | GO:0036006 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.2 | 0.9 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.2 | 4.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.2 | 1.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.2 | 2.9 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 0.6 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.2 | 3.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.2 | 1.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.2 | 1.4 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.2 | 1.2 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.2 | 0.8 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.2 | 1.2 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.2 | 0.6 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.2 | 0.8 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 | 0.6 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.2 | 0.9 | GO:0051189 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.2 | 0.9 | GO:1902047 | polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) |
| 0.2 | 1.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.2 | 0.6 | GO:2000983 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.2 | 0.9 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.2 | 1.8 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.2 | 0.5 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) |
| 0.2 | 0.5 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.2 | 2.6 | GO:0036065 | fucosylation(GO:0036065) |
| 0.2 | 0.9 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.2 | 1.5 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.2 | 0.5 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.2 | 2.7 | GO:1903861 | regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.2 | 7.4 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.2 | 1.0 | GO:0098543 | detection of bacterium(GO:0016045) negative regulation of natural killer cell activation(GO:0032815) detection of other organism(GO:0098543) |
| 0.2 | 1.3 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.2 | 1.3 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.2 | 0.5 | GO:0002585 | positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) |
| 0.2 | 0.3 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.2 | 1.6 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.2 | 0.8 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.2 | 2.3 | GO:0006152 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.2 | 0.3 | GO:0061043 | regulation of vascular wound healing(GO:0061043) |
| 0.2 | 1.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.2 | 0.5 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.2 | 0.3 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.2 | 1.4 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.2 | 1.4 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 1.3 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 2.9 | GO:0095500 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 1.0 | GO:0015840 | urea transport(GO:0015840) urea transmembrane transport(GO:0071918) |
| 0.1 | 0.3 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 0.8 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.1 | 0.8 | GO:1903056 | regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.1 | 0.4 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) protein K29-linked ubiquitination(GO:0035519) negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 | 0.4 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.1 | 0.9 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.4 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.1 | 0.3 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.1 | 0.6 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.1 | 0.4 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 0.5 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 0.7 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 0.5 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.6 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.1 | 2.4 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.1 | 0.4 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.1 | 0.7 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.1 | 1.3 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.1 | 1.3 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.1 | 0.4 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.1 | 2.8 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 4.1 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.1 | 1.3 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 1.8 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.1 | 1.5 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 1.0 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 1.4 | GO:2000191 | regulation of fatty acid transport(GO:2000191) |
| 0.1 | 0.8 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 0.9 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.6 | GO:1903352 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.1 | 2.4 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 1.0 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 | 0.3 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.1 | 0.6 | GO:0033005 | positive regulation of mast cell activation(GO:0033005) positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 | 1.0 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 0.5 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.1 | 1.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.9 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.1 | 0.6 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.1 | 0.4 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 1.7 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.1 | 1.2 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.1 | 0.6 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 1.7 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.5 | GO:2000505 | regulation of energy homeostasis(GO:2000505) |
| 0.1 | 0.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 1.2 | GO:0014887 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.1 | 0.2 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.1 | 4.4 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 0.5 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 1.5 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.1 | 1.3 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 2.6 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.1 | 1.2 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.1 | 0.8 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.1 | 0.3 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.1 | 3.9 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 0.2 | GO:0070589 | cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule metabolic process(GO:0044036) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) cell wall organization or biogenesis(GO:0071554) |
| 0.1 | 0.3 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.1 | 3.0 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.1 | 1.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.7 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 1.2 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.1 | 2.5 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 1.8 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.1 | 0.7 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 0.6 | GO:0097070 | ductus arteriosus closure(GO:0097070) |
| 0.1 | 0.7 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.4 | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.1 | 0.7 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.6 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 0.1 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.1 | 0.5 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.1 | 0.2 | GO:2000668 | dendritic cell apoptotic process(GO:0097048) regulation of dendritic cell apoptotic process(GO:2000668) |
| 0.1 | 1.9 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 1.0 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.1 | 0.7 | GO:0010543 | regulation of platelet activation(GO:0010543) |
| 0.1 | 0.2 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 0.5 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.9 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.6 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 0.5 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 0.5 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.1 | 0.5 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.7 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.1 | 0.2 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.1 | 0.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.2 | GO:2000809 | positive regulation of presynaptic membrane organization(GO:1901631) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 | 1.1 | GO:0051930 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.1 | 0.4 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.1 | 0.4 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 0.5 | GO:0071225 | cellular response to muramyl dipeptide(GO:0071225) |
| 0.1 | 1.7 | GO:0046889 | positive regulation of lipid biosynthetic process(GO:0046889) |
| 0.1 | 0.5 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.1 | 0.3 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.1 | 0.3 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.1 | 0.7 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.1 | 0.4 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 3.1 | GO:0048709 | oligodendrocyte differentiation(GO:0048709) |
| 0.1 | 2.0 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.2 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 3.4 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 3.6 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.0 | 0.3 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.2 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 3.1 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 0.7 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) kynurenine metabolic process(GO:0070189) |
| 0.0 | 0.6 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.4 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.3 | GO:0046621 | negative regulation of organ growth(GO:0046621) |
| 0.0 | 2.1 | GO:0031960 | response to corticosteroid(GO:0031960) response to glucocorticoid(GO:0051384) |
| 0.0 | 0.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 1.5 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.2 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 2.6 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 2.1 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.2 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.6 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 | 0.6 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.6 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.0 | 0.9 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 2.7 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 1.3 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 1.0 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 1.1 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 1.0 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.0 | 0.5 | GO:0060343 | trabecula formation(GO:0060343) |
| 0.0 | 1.1 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.6 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 1.4 | GO:0045806 | negative regulation of endocytosis(GO:0045806) |
| 0.0 | 0.5 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.3 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.0 | 0.2 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.1 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 0.0 | 0.6 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 1.0 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.3 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.4 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.4 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.4 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.0 | 0.7 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.5 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.6 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 3.9 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 1.1 | GO:0001658 | branching involved in ureteric bud morphogenesis(GO:0001658) |
| 0.0 | 1.5 | GO:0006661 | phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.0 | 0.3 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 0.6 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.0 | 0.2 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.6 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.8 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 1.0 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.1 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.6 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.2 | GO:0044782 | cilium organization(GO:0044782) |
| 0.0 | 0.4 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0097049 | motor neuron apoptotic process(GO:0097049) regulation of motor neuron apoptotic process(GO:2000671) |
| 0.0 | 0.1 | GO:0035886 | vascular smooth muscle cell differentiation(GO:0035886) |
| 0.0 | 0.7 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.3 | GO:0014044 | Schwann cell development(GO:0014044) myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.4 | GO:0014904 | myotube cell development(GO:0014904) |
| 0.0 | 1.4 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.7 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.2 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 1.3 | GO:0030832 | regulation of actin filament length(GO:0030832) |
| 0.0 | 0.7 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.0 | 0.1 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.4 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.8 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 0.5 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.2 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.2 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.4 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.0 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.2 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.1 | GO:0034612 | response to tumor necrosis factor(GO:0034612) |
| 0.0 | 0.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 13.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 1.7 | 8.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 1.5 | 7.7 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 1.4 | 5.7 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.9 | 4.7 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.9 | 4.6 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.9 | 2.6 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.8 | 2.5 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.8 | 4.7 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.7 | 3.4 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.7 | 3.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.6 | 8.5 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.5 | 4.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.5 | 4.4 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.5 | 4.7 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.5 | 1.8 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.5 | 1.4 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.4 | 1.3 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.4 | 1.2 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.4 | 2.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.4 | 1.8 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.4 | 6.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.3 | 11.5 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.3 | 3.4 | GO:0031091 | platelet alpha granule(GO:0031091) extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.3 | 2.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.3 | 3.6 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.3 | 0.8 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.3 | 1.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.3 | 1.1 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.3 | 3.4 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.3 | 2.6 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.2 | 2.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.2 | 2.6 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.2 | 1.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.2 | 3.6 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.2 | 7.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.2 | 4.3 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.2 | 0.6 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.2 | 2.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 1.0 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.2 | 2.2 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.2 | 2.8 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.2 | 5.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 0.5 | GO:0031251 | PAN complex(GO:0031251) |
| 0.2 | 2.4 | GO:0043196 | varicosity(GO:0043196) |
| 0.2 | 6.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 0.5 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.2 | 0.8 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.2 | 0.8 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.2 | 1.6 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 2.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 1.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.9 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 1.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 2.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 5.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 2.9 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.1 | 1.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 1.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.0 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 0.4 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.1 | 0.5 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.1 | 1.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.9 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 2.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 1.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.3 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 0.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.8 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 1.5 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 0.3 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.1 | 3.0 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.3 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.5 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 3.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.5 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 1.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.9 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 0.6 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 1.7 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 1.7 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.2 | GO:0005940 | septin ring(GO:0005940) |
| 0.1 | 0.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 2.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.7 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 4.7 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 0.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.1 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 0.6 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 0.6 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 2.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.4 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.1 | 0.9 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 14.6 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 1.0 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 2.0 | GO:0030117 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.1 | 0.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 1.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 0.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.5 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.8 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.2 | GO:0031502 | dolichyl-phosphate-mannose-protein mannosyltransferase complex(GO:0031502) |
| 0.1 | 0.4 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 3.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.9 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 2.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 9.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 1.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 1.8 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 1.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 2.6 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.0 | 8.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 2.7 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 3.3 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 3.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.5 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 2.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.0 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.8 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 1.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0043657 | host(GO:0018995) host cell part(GO:0033643) host cell(GO:0043657) |
| 0.0 | 1.4 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.1 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.0 | 1.3 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 1.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 2.4 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 8.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.7 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 2.1 | 6.3 | GO:0004368 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) |
| 1.6 | 8.1 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 1.4 | 4.3 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 1.4 | 4.1 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 1.4 | 13.6 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 1.3 | 3.9 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 1.1 | 2.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 1.0 | 3.0 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 1.0 | 4.9 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.9 | 2.7 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.9 | 3.6 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.8 | 2.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.8 | 3.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.8 | 5.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.7 | 2.7 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.7 | 2.0 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.6 | 8.6 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.6 | 3.6 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.6 | 1.8 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.6 | 3.4 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.6 | 2.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.5 | 2.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.5 | 4.7 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.5 | 4.7 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.5 | 3.7 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.5 | 0.5 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.5 | 6.9 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.5 | 2.4 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.5 | 4.7 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.5 | 2.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.5 | 10.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.5 | 3.6 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.4 | 1.3 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.4 | 2.9 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.4 | 2.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.4 | 2.8 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.4 | 2.0 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.4 | 2.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.4 | 3.8 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.4 | 1.5 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.4 | 1.8 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.4 | 1.8 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.4 | 2.9 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.4 | 2.6 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.4 | 2.5 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.3 | 3.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.3 | 3.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.3 | 1.0 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.3 | 0.3 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.3 | 0.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.3 | 1.8 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.3 | 1.5 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.3 | 1.2 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.3 | 1.7 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.3 | 1.7 | GO:0009374 | biotin binding(GO:0009374) |
| 0.3 | 3.5 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.3 | 1.7 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.3 | 4.8 | GO:0042805 | actinin binding(GO:0042805) |
| 0.3 | 1.7 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.3 | 6.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.3 | 1.6 | GO:0015108 | chloride transmembrane transporter activity(GO:0015108) |
| 0.3 | 2.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.3 | 1.6 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.3 | 0.8 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.3 | 0.8 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.2 | 1.0 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.2 | 1.5 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.2 | 1.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.2 | 2.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 0.7 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.2 | 2.3 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.2 | 1.7 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.2 | 6.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 1.8 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.2 | 1.2 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.2 | 2.2 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.2 | 0.6 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.2 | 1.0 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.2 | 0.9 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.2 | 1.7 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.2 | 1.4 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.2 | 0.7 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.2 | 1.9 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.2 | 1.0 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.2 | 1.0 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 0.5 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.2 | 0.8 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.2 | 1.0 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.2 | 0.8 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.2 | 0.7 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.2 | 0.8 | GO:0051880 | Y-form DNA binding(GO:0000403) bubble DNA binding(GO:0000405) G-quadruplex DNA binding(GO:0051880) |
| 0.2 | 1.6 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.2 | 2.9 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 3.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.2 | 0.3 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.2 | 2.6 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.2 | 1.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.1 | 4.0 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 1.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 1.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 0.6 | GO:0004954 | icosanoid receptor activity(GO:0004953) prostanoid receptor activity(GO:0004954) prostaglandin receptor activity(GO:0004955) |
| 0.1 | 0.9 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 1.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 2.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.5 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin binding(GO:0051378) serotonin receptor activity(GO:0099589) |
| 0.1 | 3.6 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.1 | 2.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 3.3 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 2.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 1.9 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.2 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.1 | 0.7 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 1.0 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.8 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.1 | 0.8 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.5 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.1 | 0.9 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.1 | 4.4 | GO:0004120 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.1 | 0.3 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.1 | 1.7 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.8 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 0.9 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 0.6 | GO:0034916 | 4-methyloctanoyl-CoA dehydrogenase activity(GO:0034580) naphthyl-2-methyl-succinyl-CoA dehydrogenase activity(GO:0034845) 2-methylhexanoyl-CoA dehydrogenase activity(GO:0034916) propionyl-CoA dehydrogenase activity(GO:0043820) thiol-driven fumarate reductase activity(GO:0043830) coenzyme F420-dependent 2,4,6-trinitrophenol reductase activity(GO:0052758) coenzyme F420-dependent 2,4,6-trinitrophenol hydride reductase activity(GO:0052759) coenzyme F420-dependent 2,4-dinitrophenol reductase activity(GO:0052760) |
| 0.1 | 1.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.8 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 3.9 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 2.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.8 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 0.8 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) |
| 0.1 | 1.7 | GO:0050253 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) retinyl-palmitate esterase activity(GO:0050253) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.1 | 1.0 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 2.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 3.7 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 14.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 4.0 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 0.5 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 0.6 | GO:0015189 | L-ornithine transmembrane transporter activity(GO:0000064) arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 1.0 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.9 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 0.5 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 3.8 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 1.2 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 4.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 3.6 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 1.2 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 0.1 | 1.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.5 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.1 | 1.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.7 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.7 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 2.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.6 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.1 | 0.8 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 0.6 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.1 | 1.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 3.2 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 0.8 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 1.1 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 0.7 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 4.1 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 0.2 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 0.5 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.2 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.1 | 0.4 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 0.3 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.1 | 0.6 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.6 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.4 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 1.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 4.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0015173 | hydrogen:amino acid symporter activity(GO:0005280) aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 1.9 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.6 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 1.7 | GO:0018602 | sulfonate dioxygenase activity(GO:0000907) 2,4-dichlorophenoxyacetate alpha-ketoglutarate dioxygenase activity(GO:0018602) hypophosphite dioxygenase activity(GO:0034792) gibberellin 2-beta-dioxygenase activity(GO:0045543) C-19 gibberellin 2-beta-dioxygenase activity(GO:0052634) C-20 gibberellin 2-beta-dioxygenase activity(GO:0052635) |
| 0.0 | 0.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 1.0 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 1.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.3 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 1.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 1.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.6 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.8 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.2 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 1.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 1.2 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 1.0 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 1.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.8 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.0 | 0.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 3.5 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.1 | GO:0045127 | N-acylmannosamine kinase activity(GO:0009384) N-acetylglucosamine kinase activity(GO:0045127) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.5 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.3 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.3 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.4 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 1.2 | GO:0008748 | N-ethylmaleimide reductase activity(GO:0008748) reduced coenzyme F420 dehydrogenase activity(GO:0043738) sulfur oxygenase reductase activity(GO:0043826) malolactic enzyme activity(GO:0043883) epoxyqueuosine reductase activity(GO:0052693) |
| 0.0 | 2.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.0 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.3 | GO:0016443 | bidentate ribonuclease III activity(GO:0016443) |
| 0.0 | 0.5 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.0 | 0.4 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 1.9 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.5 | GO:0034946 | 2-oxoglutaryl-CoA thioesterase activity(GO:0034843) 2,4,4-trimethyl-3-oxopentanoyl-CoA thioesterase activity(GO:0034869) 3-isopropylbut-3-enoyl-CoA thioesterase activity(GO:0034946) glutaryl-CoA hydrolase activity(GO:0044466) |
| 0.0 | 0.2 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 11.9 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 2.8 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.0 | 0.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.4 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.4 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.1 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.0 | 0.2 | GO:0070636 | purine-specific mismatch base pair DNA N-glycosylase activity(GO:0000701) single-strand selective uracil DNA N-glycosylase activity(GO:0017065) nicotinamide riboside hydrolase activity(GO:0070635) nicotinic acid riboside hydrolase activity(GO:0070636) deoxyribonucleoside 5'-monophosphate N-glycosidase activity(GO:0070694) |
| 0.0 | 0.5 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.7 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 1.7 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.2 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 1.8 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.0 | 2.5 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.5 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.0 | 1.7 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.6 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 1.0 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 1.3 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.0 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.8 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 2.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.2 | GO:0019887 | protein kinase regulator activity(GO:0019887) |