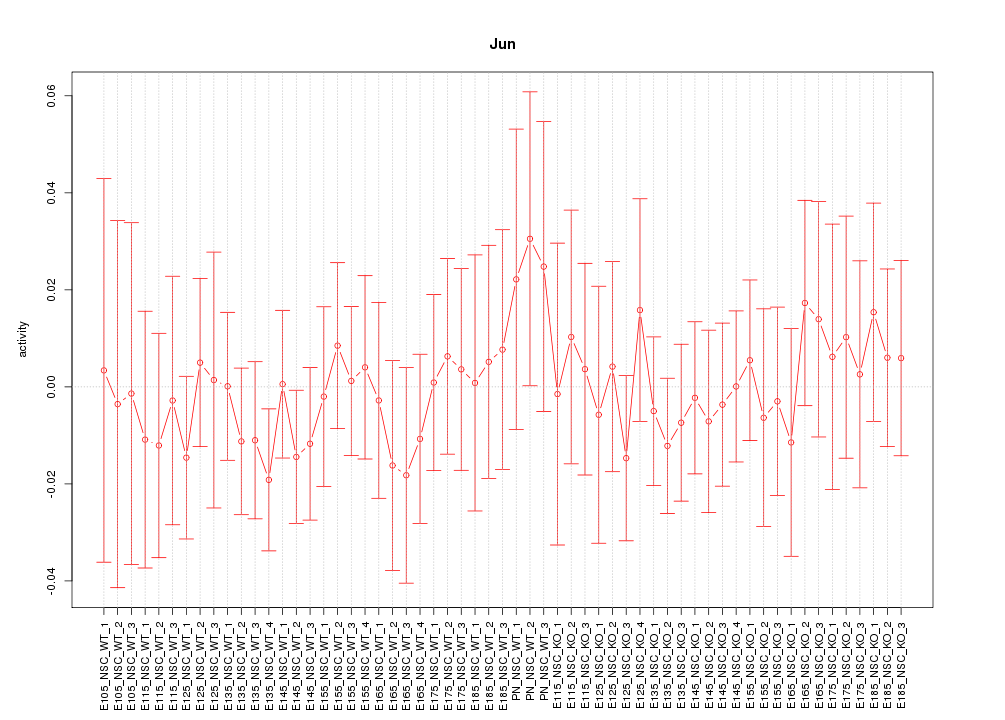
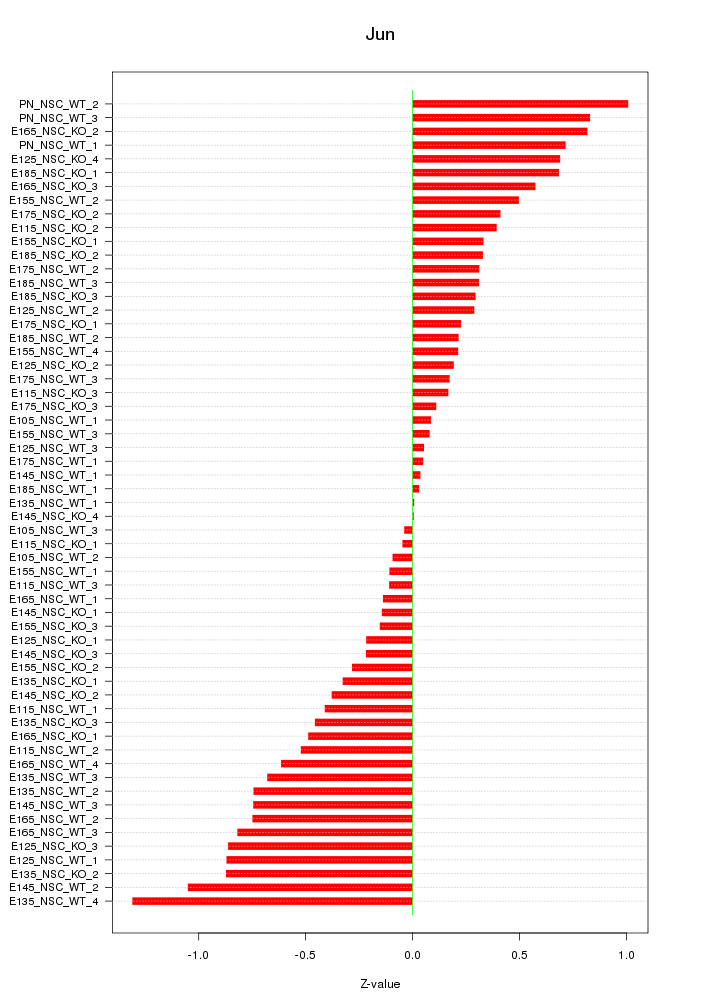
Motif ID: Jun
Z-value: 0.508
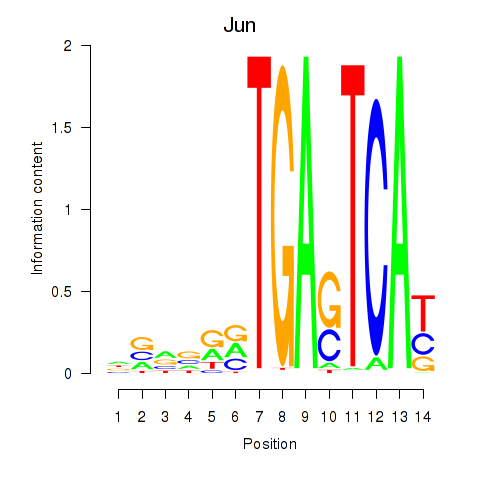
Transcription factors associated with Jun:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Jun | ENSMUSG00000052684.3 | Jun |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Jun | mm10_v2_chr4_-_95052188_95052222 | 0.39 | 2.5e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 6.8 | GO:1902847 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) |
| 0.8 | 2.5 | GO:0043465 | regulation of fermentation(GO:0043465) negative regulation of fermentation(GO:1901003) |
| 0.7 | 5.9 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.7 | 2.9 | GO:1904451 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.7 | 2.7 | GO:1904177 | regulation of adipose tissue development(GO:1904177) |
| 0.6 | 3.2 | GO:0060161 | positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.6 | 1.7 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 0.4 | 1.7 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.4 | 1.2 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.4 | 1.4 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.4 | 1.8 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.3 | 2.7 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.3 | 1.3 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.3 | 0.9 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.3 | 3.0 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.2 | 1.0 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.2 | 1.0 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) cellular response to copper ion(GO:0071280) |
| 0.2 | 0.7 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) regulation of corticosterone secretion(GO:2000852) |
| 0.2 | 1.0 | GO:2000587 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.2 | 1.0 | GO:0046133 | pyrimidine ribonucleoside catabolic process(GO:0046133) |
| 0.2 | 0.6 | GO:0048388 | endosomal lumen acidification(GO:0048388) synaptic vesicle lumen acidification(GO:0097401) |
| 0.2 | 0.8 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.2 | 0.7 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.2 | 1.3 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.2 | 1.2 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.2 | 0.5 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.2 | 0.5 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.1 | 1.8 | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) |
| 0.1 | 0.6 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.1 | 0.7 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.1 | 0.7 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 1.6 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 0.4 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) |
| 0.1 | 0.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 1.2 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 0.4 | GO:0042560 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.1 | 0.8 | GO:0051036 | regulation of endosome size(GO:0051036) receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 1.3 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.1 | 1.3 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.7 | GO:1900451 | positive regulation of glutamate receptor signaling pathway(GO:1900451) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.1 | 0.7 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 | 0.6 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.1 | 0.5 | GO:0032911 | nerve growth factor production(GO:0032902) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 | 2.0 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.1 | 0.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.5 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) positive regulation of receptor binding(GO:1900122) |
| 0.1 | 0.8 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 0.3 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) regulation of the force of heart contraction by chemical signal(GO:0003057) |
| 0.1 | 2.4 | GO:0034113 | heterotypic cell-cell adhesion(GO:0034113) |
| 0.1 | 0.9 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.1 | 0.9 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 | 0.4 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.1 | 0.4 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.1 | 0.5 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.1 | 0.5 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 1.1 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 1.4 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.2 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 1.4 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 0.9 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.1 | 0.7 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 1.4 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.1 | 0.3 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.1 | 1.0 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.9 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.1 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.0 | 2.1 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.2 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.3 | GO:1902805 | positive regulation of synaptic vesicle transport(GO:1902805) |
| 0.0 | 0.3 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.4 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.0 | 0.1 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 0.2 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.2 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.8 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.2 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.4 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.6 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 1.2 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.4 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.5 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.3 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.4 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.0 | GO:0042275 | error-free postreplication DNA repair(GO:0042275) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 6.8 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) neurofibrillary tangle(GO:0097418) |
| 0.8 | 2.4 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.7 | 2.9 | GO:1990795 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) |
| 0.4 | 2.7 | GO:0005638 | lamin filament(GO:0005638) |
| 0.3 | 0.3 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.3 | 1.2 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.2 | 1.4 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.2 | 2.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.2 | 3.2 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 0.9 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.1 | 3.2 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 1.3 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 1.4 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 1.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 0.5 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 0.3 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 0.8 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 0.3 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.1 | 1.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 3.6 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 2.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.4 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 2.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 3.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.0 | 2.3 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 4.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.0 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 1.0 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 4.6 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.9 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.6 | 1.8 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) |
| 0.4 | 2.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.4 | 2.7 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.4 | 2.9 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.3 | 1.0 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.3 | 1.7 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.3 | 1.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.3 | 1.3 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.3 | 0.9 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.3 | 6.8 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.3 | 1.7 | GO:0070728 | leucine binding(GO:0070728) |
| 0.3 | 1.3 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.2 | 1.5 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.2 | 1.4 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.2 | 0.9 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.2 | 0.7 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.1 | 0.6 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 3.2 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.1 | 1.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.4 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.1 | 1.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 1.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 2.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 2.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.3 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) |
| 0.1 | 1.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 2.4 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 0.8 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.1 | 1.3 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 0.2 | GO:0004942 | anaphylatoxin receptor activity(GO:0004942) |
| 0.1 | 0.9 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 1.0 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.1 | 0.6 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.5 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 1.0 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.8 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.3 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.5 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.6 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 0.2 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.0 | 2.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.3 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.4 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.8 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 2.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 1.8 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.9 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.2 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.5 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.5 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 1.0 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 1.1 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.3 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 1.0 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 2.2 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.4 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.6 | GO:0030551 | cyclic nucleotide binding(GO:0030551) |
| 0.0 | 0.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.9 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.3 | GO:0045296 | cadherin binding(GO:0045296) |