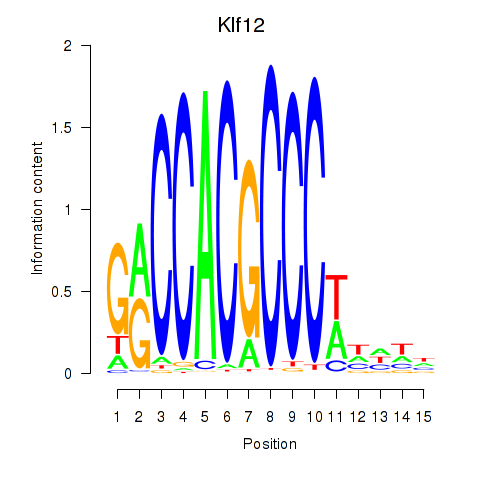
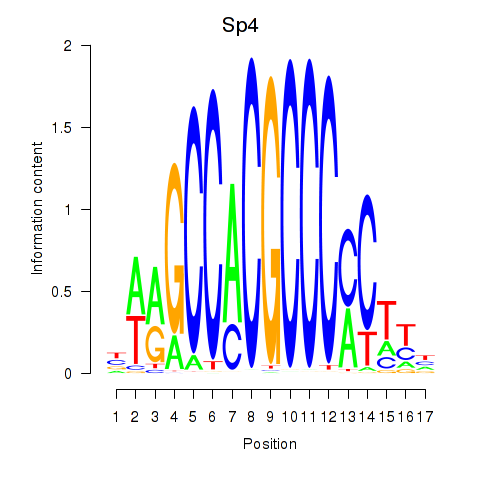
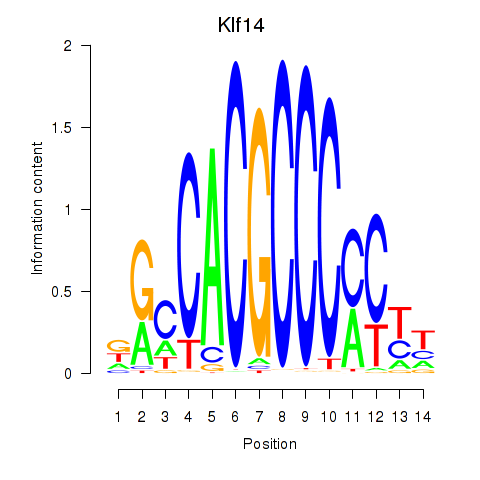
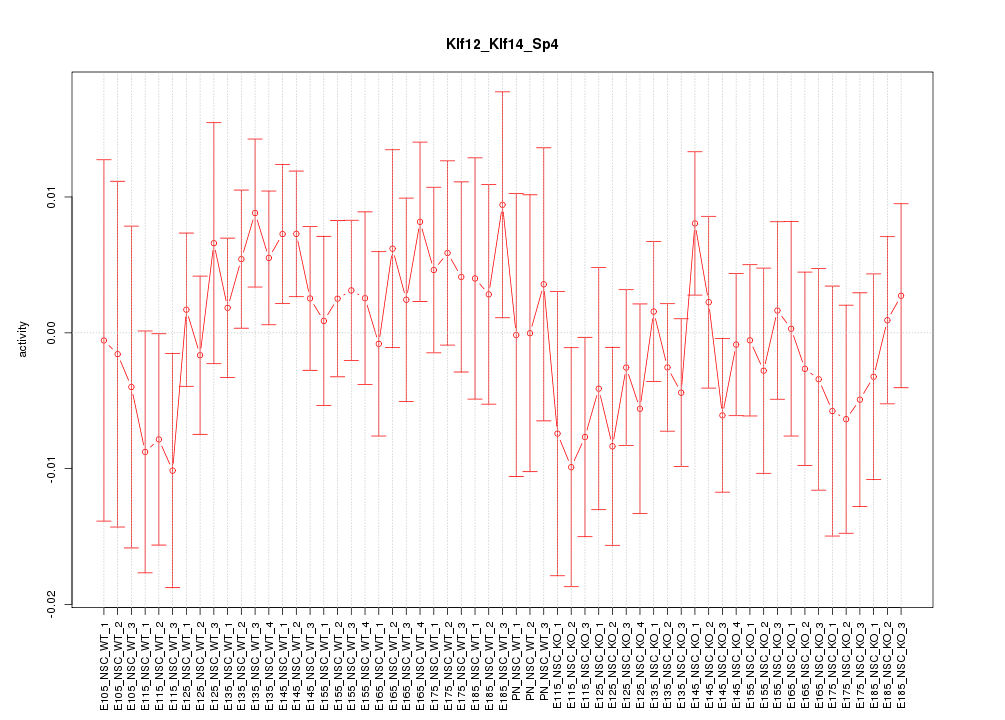
| 1.7 |
5.1 |
GO:0030827 |
negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.7 |
2.7 |
GO:0009597 |
detection of virus(GO:0009597) |
| 0.6 |
6.4 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.6 |
8.3 |
GO:0071257 |
cellular response to electrical stimulus(GO:0071257) |
| 0.6 |
1.8 |
GO:0035928 |
rRNA import into mitochondrion(GO:0035928) |
| 0.6 |
2.3 |
GO:0072344 |
rescue of stalled ribosome(GO:0072344) |
| 0.5 |
1.5 |
GO:0006578 |
amino-acid betaine biosynthetic process(GO:0006578) |
| 0.5 |
2.5 |
GO:0032485 |
regulation of Ral protein signal transduction(GO:0032485) |
| 0.5 |
1.5 |
GO:1904685 |
positive regulation of metalloendopeptidase activity(GO:1904685) |
| 0.5 |
2.4 |
GO:1904683 |
regulation of metalloendopeptidase activity(GO:1904683) |
| 0.5 |
1.4 |
GO:0043988 |
histone H3-S28 phosphorylation(GO:0043988) |
| 0.5 |
2.7 |
GO:0035743 |
CD4-positive, alpha-beta T cell cytokine production(GO:0035743) T-helper 2 cell cytokine production(GO:0035745) |
| 0.4 |
1.2 |
GO:1903061 |
regulation of protein lipidation(GO:1903059) positive regulation of protein lipidation(GO:1903061) |
| 0.4 |
1.6 |
GO:0032489 |
regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.4 |
1.6 |
GO:0000379 |
tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.4 |
1.2 |
GO:0006601 |
creatine biosynthetic process(GO:0006601) |
| 0.4 |
1.4 |
GO:0010046 |
response to mycotoxin(GO:0010046) |
| 0.4 |
1.1 |
GO:0035910 |
ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.3 |
0.3 |
GO:0032911 |
negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.3 |
1.7 |
GO:0070164 |
negative regulation of adiponectin secretion(GO:0070164) |
| 0.3 |
1.3 |
GO:2000346 |
negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.3 |
1.0 |
GO:1902071 |
regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.3 |
0.9 |
GO:0046340 |
diacylglycerol catabolic process(GO:0046340) |
| 0.3 |
1.2 |
GO:0035609 |
C-terminal protein deglutamylation(GO:0035609) |
| 0.3 |
0.3 |
GO:1902724 |
positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.3 |
0.6 |
GO:0097494 |
regulation of vesicle size(GO:0097494) |
| 0.3 |
1.2 |
GO:1901642 |
purine nucleoside transmembrane transport(GO:0015860) purine-containing compound transmembrane transport(GO:0072530) nucleoside transmembrane transport(GO:1901642) |
| 0.3 |
2.8 |
GO:0045820 |
negative regulation of glycolytic process(GO:0045820) |
| 0.3 |
1.7 |
GO:0016557 |
peroxisome membrane biogenesis(GO:0016557) |
| 0.3 |
1.1 |
GO:1901475 |
pyruvate transmembrane transport(GO:1901475) |
| 0.3 |
0.8 |
GO:0008612 |
peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.3 |
0.8 |
GO:0019482 |
beta-alanine metabolic process(GO:0019482) |
| 0.2 |
0.7 |
GO:0043379 |
memory T cell differentiation(GO:0043379) |
| 0.2 |
0.5 |
GO:1902566 |
regulation of eosinophil activation(GO:1902566) |
| 0.2 |
0.7 |
GO:0046168 |
glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.2 |
1.9 |
GO:0009181 |
purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.2 |
1.0 |
GO:0007039 |
protein catabolic process in the vacuole(GO:0007039) |
| 0.2 |
1.6 |
GO:0030382 |
sperm mitochondrion organization(GO:0030382) |
| 0.2 |
1.9 |
GO:1902669 |
positive regulation of axon extension involved in axon guidance(GO:0048842) positive regulation of axon guidance(GO:1902669) |
| 0.2 |
0.7 |
GO:0034035 |
purine ribonucleoside bisphosphate metabolic process(GO:0034035) |
| 0.2 |
0.9 |
GO:0099624 |
regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) atrial cardiac muscle cell membrane repolarization(GO:0099624) |
| 0.2 |
2.2 |
GO:0061158 |
3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.2 |
0.7 |
GO:0009298 |
GDP-mannose biosynthetic process(GO:0009298) |
| 0.2 |
1.1 |
GO:0034380 |
high-density lipoprotein particle assembly(GO:0034380) |
| 0.2 |
0.9 |
GO:0048597 |
post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.2 |
0.9 |
GO:2000048 |
negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.2 |
0.6 |
GO:0034447 |
very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.2 |
0.6 |
GO:0010533 |
regulation of activation of Janus kinase activity(GO:0010533) regulation of activation of JAK2 kinase activity(GO:0010534) |
| 0.2 |
0.8 |
GO:0070086 |
ubiquitin-dependent endocytosis(GO:0070086) |
| 0.2 |
0.8 |
GO:0042373 |
vitamin K metabolic process(GO:0042373) |
| 0.2 |
0.6 |
GO:0061144 |
alveolar secondary septum development(GO:0061144) |
| 0.2 |
0.6 |
GO:0090327 |
negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.2 |
0.6 |
GO:0006669 |
sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.2 |
0.4 |
GO:0043696 |
dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.2 |
0.6 |
GO:0018103 |
protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.2 |
0.6 |
GO:0072223 |
metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 0.2 |
0.6 |
GO:0032915 |
positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.2 |
0.4 |
GO:1904468 |
negative regulation of tumor necrosis factor secretion(GO:1904468) |
| 0.2 |
1.0 |
GO:0045541 |
negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.2 |
0.6 |
GO:0001920 |
negative regulation of receptor recycling(GO:0001920) |
| 0.2 |
0.8 |
GO:0010847 |
regulation of chromatin assembly(GO:0010847) |
| 0.2 |
0.2 |
GO:0044340 |
canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.2 |
2.3 |
GO:0019262 |
N-acetylneuraminate catabolic process(GO:0019262) |
| 0.2 |
2.1 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.2 |
0.6 |
GO:0035519 |
protein K29-linked ubiquitination(GO:0035519) |
| 0.2 |
0.7 |
GO:0097298 |
regulation of nucleus size(GO:0097298) |
| 0.2 |
0.9 |
GO:0009313 |
oligosaccharide catabolic process(GO:0009313) |
| 0.2 |
0.7 |
GO:0035934 |
corticosterone secretion(GO:0035934) |
| 0.2 |
0.9 |
GO:1902837 |
amino acid import into cell(GO:1902837) |
| 0.2 |
1.1 |
GO:0032804 |
negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 0.2 |
0.2 |
GO:0007113 |
endomitotic cell cycle(GO:0007113) |
| 0.2 |
0.5 |
GO:0060468 |
prevention of polyspermy(GO:0060468) |
| 0.2 |
0.9 |
GO:0016127 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.2 |
0.4 |
GO:0097155 |
fasciculation of sensory neuron axon(GO:0097155) |
| 0.2 |
0.7 |
GO:0007066 |
female meiosis sister chromatid cohesion(GO:0007066) |
| 0.2 |
0.3 |
GO:0036166 |
phenotypic switching(GO:0036166) |
| 0.2 |
0.8 |
GO:0010032 |
meiotic chromosome condensation(GO:0010032) |
| 0.2 |
0.8 |
GO:0010961 |
cellular magnesium ion homeostasis(GO:0010961) |
| 0.2 |
1.0 |
GO:2000786 |
positive regulation of autophagosome assembly(GO:2000786) |
| 0.2 |
0.5 |
GO:0038161 |
prolactin signaling pathway(GO:0038161) |
| 0.2 |
0.6 |
GO:0046469 |
platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.2 |
2.6 |
GO:0071378 |
growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.2 |
0.5 |
GO:0015919 |
peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.2 |
0.6 |
GO:1903026 |
negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.2 |
0.5 |
GO:0006682 |
galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 |
0.8 |
GO:1902083 |
negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.2 |
1.1 |
GO:0002591 |
positive regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002591) |
| 0.2 |
0.9 |
GO:0006384 |
transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 |
0.7 |
GO:0043691 |
reverse cholesterol transport(GO:0043691) |
| 0.1 |
0.3 |
GO:0042505 |
tyrosine phosphorylation of Stat6 protein(GO:0042505) regulation of tyrosine phosphorylation of Stat6 protein(GO:0042525) |
| 0.1 |
0.4 |
GO:0034241 |
positive regulation of macrophage fusion(GO:0034241) |
| 0.1 |
1.4 |
GO:2000051 |
negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 |
1.4 |
GO:0048199 |
vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.1 |
0.8 |
GO:0019336 |
phenol-containing compound catabolic process(GO:0019336) |
| 0.1 |
1.2 |
GO:0006527 |
arginine catabolic process(GO:0006527) |
| 0.1 |
0.7 |
GO:0018125 |
peptidyl-cysteine methylation(GO:0018125) |
| 0.1 |
0.7 |
GO:0001561 |
fatty acid alpha-oxidation(GO:0001561) |
| 0.1 |
0.7 |
GO:0072383 |
plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 |
0.5 |
GO:0034441 |
plasma lipoprotein particle oxidation(GO:0034441) |
| 0.1 |
0.5 |
GO:2000525 |
regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.1 |
0.8 |
GO:0090383 |
phagosome acidification(GO:0090383) |
| 0.1 |
0.4 |
GO:0033566 |
gamma-tubulin complex localization(GO:0033566) |
| 0.1 |
0.4 |
GO:0051342 |
regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.1 |
0.5 |
GO:0048861 |
leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 |
0.9 |
GO:0051036 |
regulation of endosome size(GO:0051036) |
| 0.1 |
0.4 |
GO:0060854 |
patterning of lymph vessels(GO:0060854) |
| 0.1 |
0.6 |
GO:0030205 |
dermatan sulfate metabolic process(GO:0030205) |
| 0.1 |
0.9 |
GO:0014816 |
skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 |
0.4 |
GO:0021933 |
radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 0.1 |
0.5 |
GO:0033540 |
fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 |
0.7 |
GO:2000047 |
regulation of cell-cell adhesion mediated by cadherin(GO:2000047) positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.1 |
0.4 |
GO:0042939 |
renal sodium ion transport(GO:0003096) glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.1 |
0.7 |
GO:0071415 |
cellular response to caffeine(GO:0071313) cellular response to purine-containing compound(GO:0071415) |
| 0.1 |
0.1 |
GO:1904467 |
regulation of tumor necrosis factor secretion(GO:1904467) tumor necrosis factor secretion(GO:1990774) |
| 0.1 |
0.2 |
GO:1902445 |
regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.1 |
0.2 |
GO:0070555 |
response to interleukin-1(GO:0070555) |
| 0.1 |
0.6 |
GO:0051697 |
protein delipidation(GO:0051697) |
| 0.1 |
1.0 |
GO:0071569 |
protein ufmylation(GO:0071569) |
| 0.1 |
0.2 |
GO:0003104 |
positive regulation of glomerular filtration(GO:0003104) negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.1 |
0.2 |
GO:2000705 |
regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 |
0.3 |
GO:0060455 |
negative regulation of gastric acid secretion(GO:0060455) |
| 0.1 |
0.3 |
GO:0036091 |
positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 |
1.9 |
GO:0006516 |
glycoprotein catabolic process(GO:0006516) |
| 0.1 |
0.3 |
GO:0042560 |
10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.1 |
0.3 |
GO:0006553 |
lysine metabolic process(GO:0006553) |
| 0.1 |
2.0 |
GO:0045653 |
negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 |
0.1 |
GO:0018214 |
peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 |
0.3 |
GO:0070682 |
proteasome regulatory particle assembly(GO:0070682) |
| 0.1 |
0.6 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.1 |
0.3 |
GO:0070863 |
positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.1 |
1.8 |
GO:0033539 |
fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 |
0.2 |
GO:0009182 |
purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) GDP metabolic process(GO:0046710) |
| 0.1 |
0.8 |
GO:0090232 |
positive regulation of spindle checkpoint(GO:0090232) |
| 0.1 |
0.2 |
GO:0007341 |
penetration of zona pellucida(GO:0007341) |
| 0.1 |
0.8 |
GO:0061687 |
detoxification of inorganic compound(GO:0061687) |
| 0.1 |
0.4 |
GO:0019372 |
lipoxygenase pathway(GO:0019372) |
| 0.1 |
0.6 |
GO:0030397 |
membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.1 |
0.4 |
GO:0042796 |
snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 |
0.4 |
GO:0070589 |
cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule metabolic process(GO:0044036) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) cell wall organization or biogenesis(GO:0071554) |
| 0.1 |
0.1 |
GO:0006701 |
progesterone biosynthetic process(GO:0006701) |
| 0.1 |
0.1 |
GO:2000562 |
negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.1 |
0.4 |
GO:0071475 |
cellular hyperosmotic salinity response(GO:0071475) |
| 0.1 |
1.5 |
GO:2000821 |
regulation of grooming behavior(GO:2000821) |
| 0.1 |
0.1 |
GO:0006501 |
C-terminal protein lipidation(GO:0006501) |
| 0.1 |
0.8 |
GO:0070475 |
rRNA base methylation(GO:0070475) |
| 0.1 |
0.2 |
GO:0070885 |
negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.1 |
0.4 |
GO:0072385 |
minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.1 |
0.3 |
GO:0019074 |
viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 |
0.3 |
GO:0051823 |
regulation of synapse structural plasticity(GO:0051823) |
| 0.1 |
0.3 |
GO:0090529 |
barrier septum assembly(GO:0000917) cell septum assembly(GO:0090529) |
| 0.1 |
0.3 |
GO:0030070 |
insulin processing(GO:0030070) |
| 0.1 |
0.3 |
GO:0006065 |
UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.1 |
0.3 |
GO:0060708 |
spongiotrophoblast differentiation(GO:0060708) |
| 0.1 |
0.3 |
GO:0046077 |
dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) UDP metabolic process(GO:0046048) dUDP metabolic process(GO:0046077) |
| 0.1 |
0.4 |
GO:0015788 |
UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.1 |
0.6 |
GO:0032534 |
regulation of microvillus assembly(GO:0032534) |
| 0.1 |
0.4 |
GO:0002457 |
T cell antigen processing and presentation(GO:0002457) |
| 0.1 |
0.6 |
GO:1903301 |
positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.1 |
0.3 |
GO:1900149 |
positive regulation of Schwann cell migration(GO:1900149) |
| 0.1 |
0.3 |
GO:0045448 |
regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 |
0.3 |
GO:0061526 |
acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) acetylcholine secretion(GO:0061526) |
| 0.1 |
0.2 |
GO:0007525 |
somatic muscle development(GO:0007525) |
| 0.1 |
0.2 |
GO:0021943 |
formation of radial glial scaffolds(GO:0021943) |
| 0.1 |
0.3 |
GO:0001907 |
killing by symbiont of host cells(GO:0001907) induction of programmed cell death(GO:0012502) disruption by symbiont of host cell(GO:0044004) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 |
0.4 |
GO:0010991 |
negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 |
0.8 |
GO:0006622 |
protein targeting to lysosome(GO:0006622) |
| 0.1 |
0.5 |
GO:0070417 |
cellular response to cold(GO:0070417) |
| 0.1 |
1.4 |
GO:2001014 |
regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 |
0.5 |
GO:0034393 |
positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.1 |
0.8 |
GO:0090557 |
establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 |
0.5 |
GO:0030913 |
paranodal junction assembly(GO:0030913) |
| 0.1 |
0.1 |
GO:0061153 |
trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 |
0.2 |
GO:0060754 |
positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.1 |
0.4 |
GO:0060665 |
regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 |
0.3 |
GO:0051771 |
negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.1 |
1.3 |
GO:0090161 |
Golgi ribbon formation(GO:0090161) |
| 0.1 |
1.0 |
GO:0021869 |
forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 |
0.4 |
GO:0070779 |
D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 |
0.3 |
GO:0006624 |
vacuolar protein processing(GO:0006624) |
| 0.1 |
0.4 |
GO:0097343 |
ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.1 |
0.7 |
GO:0060019 |
radial glial cell differentiation(GO:0060019) |
| 0.1 |
0.1 |
GO:0050651 |
dermatan sulfate proteoglycan biosynthetic process(GO:0050651) dermatan sulfate proteoglycan metabolic process(GO:0050655) |
| 0.1 |
0.6 |
GO:1900086 |
positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.1 |
0.9 |
GO:0048280 |
vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 |
1.1 |
GO:0000301 |
retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.1 |
0.6 |
GO:0035881 |
amacrine cell differentiation(GO:0035881) |
| 0.1 |
0.3 |
GO:0060161 |
positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.1 |
0.1 |
GO:0006642 |
triglyceride mobilization(GO:0006642) |
| 0.1 |
1.3 |
GO:0071470 |
cellular response to osmotic stress(GO:0071470) |
| 0.1 |
1.1 |
GO:2000096 |
positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 |
0.9 |
GO:0060213 |
regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 |
0.3 |
GO:1902268 |
negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 |
0.9 |
GO:0006614 |
SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 |
0.1 |
GO:0006549 |
isoleucine metabolic process(GO:0006549) |
| 0.1 |
0.3 |
GO:1900170 |
negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.1 |
0.3 |
GO:0045654 |
positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 |
0.3 |
GO:0034454 |
microtubule anchoring at centrosome(GO:0034454) microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.1 |
0.1 |
GO:0007632 |
visual behavior(GO:0007632) |
| 0.1 |
0.2 |
GO:0006703 |
estrogen biosynthetic process(GO:0006703) |
| 0.1 |
1.0 |
GO:0032211 |
negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.1 |
0.5 |
GO:0015791 |
polyol transport(GO:0015791) |
| 0.1 |
0.2 |
GO:0048388 |
endosomal lumen acidification(GO:0048388) |
| 0.1 |
0.3 |
GO:0060266 |
negative regulation of respiratory burst involved in inflammatory response(GO:0060266) negative regulation of respiratory burst(GO:0060268) |
| 0.1 |
0.1 |
GO:0042758 |
long-chain fatty acid catabolic process(GO:0042758) |
| 0.1 |
0.6 |
GO:0042989 |
sequestering of actin monomers(GO:0042989) |
| 0.1 |
0.8 |
GO:0060215 |
primitive hemopoiesis(GO:0060215) |
| 0.1 |
0.7 |
GO:0006995 |
cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 |
0.2 |
GO:0061743 |
motor learning(GO:0061743) |
| 0.1 |
0.2 |
GO:2000507 |
positive regulation of energy homeostasis(GO:2000507) |
| 0.1 |
2.6 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 |
0.4 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 |
0.1 |
GO:0010730 |
negative regulation of hydrogen peroxide metabolic process(GO:0010727) negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.1 |
0.6 |
GO:0032570 |
response to progesterone(GO:0032570) |
| 0.1 |
0.8 |
GO:0006750 |
glutathione biosynthetic process(GO:0006750) |
| 0.1 |
0.2 |
GO:0016598 |
protein arginylation(GO:0016598) |
| 0.1 |
0.2 |
GO:1904426 |
positive regulation of GTP binding(GO:1904426) regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.1 |
0.1 |
GO:0098908 |
regulation of neuronal action potential(GO:0098908) |
| 0.1 |
0.5 |
GO:0051561 |
positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.1 |
0.5 |
GO:0071318 |
cellular response to ATP(GO:0071318) |
| 0.1 |
0.3 |
GO:0042167 |
heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 |
0.2 |
GO:0044854 |
plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 |
0.2 |
GO:0038128 |
ERBB2 signaling pathway(GO:0038128) |
| 0.1 |
0.3 |
GO:0097264 |
self proteolysis(GO:0097264) |
| 0.1 |
0.6 |
GO:0097084 |
vascular smooth muscle cell development(GO:0097084) |
| 0.1 |
0.2 |
GO:1901740 |
negative regulation of myoblast fusion(GO:1901740) |
| 0.1 |
0.1 |
GO:0002447 |
eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil activation(GO:0043307) eosinophil degranulation(GO:0043308) |
| 0.1 |
0.2 |
GO:0030327 |
prenylated protein catabolic process(GO:0030327) |
| 0.1 |
0.2 |
GO:0030431 |
sleep(GO:0030431) |
| 0.1 |
0.4 |
GO:0016480 |
negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 |
0.2 |
GO:0019085 |
early viral transcription(GO:0019085) |
| 0.1 |
0.8 |
GO:0002115 |
store-operated calcium entry(GO:0002115) |
| 0.1 |
0.2 |
GO:0044314 |
protein K27-linked ubiquitination(GO:0044314) |
| 0.1 |
0.2 |
GO:0042117 |
monocyte activation(GO:0042117) |
| 0.1 |
0.2 |
GO:0042196 |
dichloromethane metabolic process(GO:0018900) chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.1 |
0.1 |
GO:0043652 |
engulfment of apoptotic cell(GO:0043652) |
| 0.1 |
0.3 |
GO:0032815 |
negative regulation of natural killer cell activation(GO:0032815) |
| 0.1 |
0.1 |
GO:0045163 |
clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 |
0.2 |
GO:0021564 |
glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 0.1 |
0.2 |
GO:0036258 |
multivesicular body assembly(GO:0036258) |
| 0.1 |
0.1 |
GO:0006971 |
hypotonic response(GO:0006971) |
| 0.1 |
0.3 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.1 |
0.4 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 |
0.2 |
GO:0019255 |
glucose 1-phosphate metabolic process(GO:0019255) |
| 0.1 |
0.2 |
GO:0021586 |
pons maturation(GO:0021586) |
| 0.1 |
0.6 |
GO:0045792 |
negative regulation of cell size(GO:0045792) |
| 0.1 |
0.2 |
GO:0010724 |
regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 |
0.6 |
GO:0090140 |
regulation of mitochondrial fission(GO:0090140) |
| 0.1 |
0.3 |
GO:0071955 |
recycling endosome to Golgi transport(GO:0071955) |
| 0.1 |
0.5 |
GO:0060363 |
cranial suture morphogenesis(GO:0060363) |
| 0.1 |
0.2 |
GO:0035990 |
tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.1 |
0.3 |
GO:0045842 |
positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 |
1.7 |
GO:0007218 |
neuropeptide signaling pathway(GO:0007218) |
| 0.0 |
0.2 |
GO:0007253 |
cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 |
0.2 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 |
0.1 |
GO:1903279 |
regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 |
0.2 |
GO:0090219 |
negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 |
0.2 |
GO:0002326 |
B cell lineage commitment(GO:0002326) |
| 0.0 |
0.0 |
GO:0031284 |
positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 |
0.2 |
GO:0006049 |
UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 |
0.3 |
GO:1901524 |
regulation of macromitophagy(GO:1901524) negative regulation of macromitophagy(GO:1901525) |
| 0.0 |
0.2 |
GO:0097012 |
response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 |
0.4 |
GO:0060837 |
blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 |
0.3 |
GO:0015862 |
uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 |
0.1 |
GO:1904431 |
positive regulation of t-circle formation(GO:1904431) |
| 0.0 |
0.4 |
GO:0003062 |
regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 |
0.1 |
GO:0043651 |
linoleic acid metabolic process(GO:0043651) |
| 0.0 |
0.0 |
GO:0090194 |
negative regulation of glomerular mesangial cell proliferation(GO:0072125) negative regulation of glomerulus development(GO:0090194) |
| 0.0 |
0.7 |
GO:0032957 |
inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 |
0.3 |
GO:0006729 |
tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 |
0.5 |
GO:0006851 |
mitochondrial calcium ion transport(GO:0006851) |
| 0.0 |
0.2 |
GO:0071985 |
multivesicular body sorting pathway(GO:0071985) |
| 0.0 |
0.1 |
GO:0033088 |
negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 |
0.2 |
GO:1900122 |
positive regulation of receptor binding(GO:1900122) |
| 0.0 |
0.3 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.0 |
0.7 |
GO:0051894 |
positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 |
0.0 |
GO:0034047 |
regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.0 |
0.1 |
GO:0051798 |
positive regulation of hair follicle development(GO:0051798) |
| 0.0 |
0.2 |
GO:0003150 |
muscular septum morphogenesis(GO:0003150) |
| 0.0 |
0.3 |
GO:0046292 |
formaldehyde metabolic process(GO:0046292) |
| 0.0 |
0.8 |
GO:0007176 |
regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.0 |
0.1 |
GO:1901029 |
negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 |
0.1 |
GO:0001306 |
age-dependent response to oxidative stress(GO:0001306) detection of oxygen(GO:0003032) age-dependent general metabolic decline(GO:0007571) |
| 0.0 |
0.2 |
GO:0006290 |
pyrimidine dimer repair(GO:0006290) |
| 0.0 |
0.2 |
GO:0006975 |
DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 |
0.0 |
GO:1901187 |
regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 |
0.5 |
GO:0035493 |
SNARE complex assembly(GO:0035493) |
| 0.0 |
0.9 |
GO:0003009 |
skeletal muscle contraction(GO:0003009) |
| 0.0 |
1.2 |
GO:0046854 |
phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 |
0.2 |
GO:0051639 |
actin filament network formation(GO:0051639) |
| 0.0 |
0.2 |
GO:0002347 |
response to tumor cell(GO:0002347) |
| 0.0 |
0.2 |
GO:0033210 |
leptin-mediated signaling pathway(GO:0033210) |
| 0.0 |
0.4 |
GO:0033008 |
positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 |
0.2 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.0 |
1.2 |
GO:0031146 |
SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 |
0.3 |
GO:2000394 |
positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 |
0.5 |
GO:0007095 |
mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 |
0.3 |
GO:0032876 |
negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 |
0.1 |
GO:0006895 |
Golgi to endosome transport(GO:0006895) |
| 0.0 |
0.2 |
GO:0003383 |
apical constriction(GO:0003383) |
| 0.0 |
0.4 |
GO:0070861 |
regulation of protein exit from endoplasmic reticulum(GO:0070861) |
| 0.0 |
0.3 |
GO:0098532 |
histone H3-K27 trimethylation(GO:0098532) |
| 0.0 |
0.2 |
GO:0009052 |
pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 |
0.0 |
GO:0046716 |
muscle cell cellular homeostasis(GO:0046716) |
| 0.0 |
0.3 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.0 |
0.2 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.0 |
0.2 |
GO:0007191 |
adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 |
0.2 |
GO:0090074 |
negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 |
0.5 |
GO:0006264 |
mitochondrial DNA replication(GO:0006264) |
| 0.0 |
0.4 |
GO:0090084 |
negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 |
0.2 |
GO:0045607 |
regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 |
0.3 |
GO:0035457 |
cellular response to interferon-alpha(GO:0035457) |
| 0.0 |
0.1 |
GO:0006538 |
glutamate catabolic process(GO:0006538) |
| 0.0 |
0.1 |
GO:0000350 |
generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 |
0.1 |
GO:0010890 |
positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 |
0.1 |
GO:1902174 |
positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 |
0.1 |
GO:0019254 |
carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 |
0.3 |
GO:0061635 |
regulation of protein complex stability(GO:0061635) |
| 0.0 |
0.3 |
GO:0021819 |
layer formation in cerebral cortex(GO:0021819) |
| 0.0 |
0.2 |
GO:0071712 |
ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 |
2.2 |
GO:0045669 |
positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 |
0.3 |
GO:0060770 |
negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 |
0.3 |
GO:0035330 |
regulation of hippo signaling(GO:0035330) |
| 0.0 |
0.4 |
GO:0061014 |
positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 |
0.2 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.0 |
0.1 |
GO:0042732 |
D-xylose metabolic process(GO:0042732) |
| 0.0 |
0.2 |
GO:0045671 |
negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 |
0.2 |
GO:0031547 |
brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 |
0.1 |
GO:2000812 |
regulation of barbed-end actin filament capping(GO:2000812) |
| 0.0 |
0.3 |
GO:0007220 |
Notch receptor processing(GO:0007220) |
| 0.0 |
0.1 |
GO:0021812 |
neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) lateral motor column neuron migration(GO:0097477) |
| 0.0 |
0.1 |
GO:0070886 |
positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 |
0.2 |
GO:0048102 |
autophagic cell death(GO:0048102) |
| 0.0 |
0.8 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
0.3 |
GO:0035635 |
entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 |
0.5 |
GO:0035518 |
histone H2A monoubiquitination(GO:0035518) |
| 0.0 |
0.4 |
GO:1902459 |
positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 |
0.2 |
GO:2000672 |
negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 |
0.2 |
GO:0021814 |
cell motility involved in cerebral cortex radial glia guided migration(GO:0021814) |
| 0.0 |
0.1 |
GO:0045078 |
positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 |
0.1 |
GO:0070317 |
negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 |
0.2 |
GO:0042866 |
pyruvate biosynthetic process(GO:0042866) |
| 0.0 |
0.2 |
GO:0006451 |
selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 |
0.1 |
GO:0070375 |
ERK5 cascade(GO:0070375) |
| 0.0 |
0.1 |
GO:0060710 |
chorio-allantoic fusion(GO:0060710) |
| 0.0 |
1.1 |
GO:0031529 |
ruffle organization(GO:0031529) |
| 0.0 |
0.4 |
GO:0031116 |
positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 |
0.1 |
GO:0001827 |
inner cell mass cell fate commitment(GO:0001827) |
| 0.0 |
0.2 |
GO:2001275 |
positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 |
0.3 |
GO:0060628 |
regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 |
0.1 |
GO:0043032 |
positive regulation of macrophage activation(GO:0043032) |
| 0.0 |
0.3 |
GO:0036120 |
cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 |
1.4 |
GO:0006635 |
fatty acid beta-oxidation(GO:0006635) |
| 0.0 |
0.1 |
GO:2000314 |
negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.0 |
0.2 |
GO:0046541 |
saliva secretion(GO:0046541) |
| 0.0 |
0.1 |
GO:0097527 |
necroptotic signaling pathway(GO:0097527) |
| 0.0 |
0.1 |
GO:0010756 |
positive regulation of plasminogen activation(GO:0010756) |
| 0.0 |
0.1 |
GO:0090160 |
Golgi to lysosome transport(GO:0090160) |
| 0.0 |
0.2 |
GO:0002315 |
marginal zone B cell differentiation(GO:0002315) |
| 0.0 |
0.1 |
GO:0046726 |
positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 |
0.2 |
GO:0015808 |
L-alanine transport(GO:0015808) |
| 0.0 |
0.3 |
GO:0070986 |
left/right axis specification(GO:0070986) |
| 0.0 |
0.2 |
GO:0015879 |
carnitine transport(GO:0015879) |
| 0.0 |
0.1 |
GO:0090148 |
membrane fission(GO:0090148) |
| 0.0 |
0.4 |
GO:0006465 |
signal peptide processing(GO:0006465) |
| 0.0 |
0.3 |
GO:0002446 |
neutrophil mediated immunity(GO:0002446) |
| 0.0 |
0.1 |
GO:0060295 |
regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 |
0.1 |
GO:0010728 |
regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.0 |
0.1 |
GO:2000074 |
regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 |
0.1 |
GO:0006659 |
phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 |
0.3 |
GO:0000920 |
cell separation after cytokinesis(GO:0000920) |
| 0.0 |
0.1 |
GO:0098501 |
polynucleotide dephosphorylation(GO:0098501) |
| 0.0 |
0.2 |
GO:1902231 |
positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 |
0.3 |
GO:0052646 |
alditol phosphate metabolic process(GO:0052646) |
| 0.0 |
0.2 |
GO:0032516 |
positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 |
0.2 |
GO:0071340 |
skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 |
1.6 |
GO:0051865 |
protein autoubiquitination(GO:0051865) |
| 0.0 |
0.2 |
GO:0030206 |
chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 |
0.3 |
GO:0042044 |
fluid transport(GO:0042044) |
| 0.0 |
0.2 |
GO:0000132 |
establishment of mitotic spindle orientation(GO:0000132) establishment of mitotic spindle localization(GO:0040001) |
| 0.0 |
0.3 |
GO:0046036 |
GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.0 |
0.9 |
GO:0006730 |
one-carbon metabolic process(GO:0006730) |
| 0.0 |
0.0 |
GO:0048227 |
plasma membrane to endosome transport(GO:0048227) |
| 0.0 |
0.3 |
GO:0034501 |
protein localization to kinetochore(GO:0034501) |
| 0.0 |
0.1 |
GO:0060216 |
definitive hemopoiesis(GO:0060216) |
| 0.0 |
0.4 |
GO:0007035 |
vacuolar acidification(GO:0007035) |
| 0.0 |
0.1 |
GO:2000015 |
regulation of determination of dorsal identity(GO:2000015) |
| 0.0 |
0.5 |
GO:0031115 |
negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 |
0.5 |
GO:0000281 |
mitotic cytokinesis(GO:0000281) |
| 0.0 |
0.1 |
GO:0006432 |
phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 |
0.2 |
GO:0031053 |
primary miRNA processing(GO:0031053) |
| 0.0 |
0.1 |
GO:0033136 |
serine phosphorylation of STAT3 protein(GO:0033136) serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 |
0.2 |
GO:0061469 |
regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 |
0.4 |
GO:0007143 |
female meiotic division(GO:0007143) |
| 0.0 |
0.0 |
GO:1901626 |
regulation of postsynaptic membrane organization(GO:1901626) |
| 0.0 |
0.1 |
GO:0070669 |
response to interleukin-2(GO:0070669) |
| 0.0 |
0.1 |
GO:0019852 |
L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 |
0.0 |
GO:0042448 |
progesterone metabolic process(GO:0042448) |
| 0.0 |
0.5 |
GO:0060074 |
synapse maturation(GO:0060074) |
| 0.0 |
0.2 |
GO:0033234 |
negative regulation of protein sumoylation(GO:0033234) |
| 0.0 |
0.0 |
GO:0006681 |
galactosylceramide metabolic process(GO:0006681) galactolipid metabolic process(GO:0019374) |
| 0.0 |
0.0 |
GO:0010701 |
positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 |
0.1 |
GO:0042126 |
nitrate metabolic process(GO:0042126) |
| 0.0 |
0.1 |
GO:0060431 |
primary lung bud formation(GO:0060431) |
| 0.0 |
1.6 |
GO:0050885 |
neuromuscular process controlling balance(GO:0050885) |
| 0.0 |
0.1 |
GO:2000807 |
regulation of synaptic vesicle clustering(GO:2000807) |
| 0.0 |
0.5 |
GO:0048026 |
positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 |
0.0 |
GO:1902410 |
mitotic cytokinetic process(GO:1902410) |
| 0.0 |
0.1 |
GO:0034316 |
negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 |
0.8 |
GO:0045600 |
positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 |
1.0 |
GO:0048843 |
negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.0 |
0.1 |
GO:0010792 |
DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 |
0.2 |
GO:0090286 |
cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 |
0.1 |
GO:2001185 |
regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.0 |
0.3 |
GO:0048266 |
behavioral response to pain(GO:0048266) |
| 0.0 |
0.3 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.0 |
0.2 |
GO:0002361 |
CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 |
0.3 |
GO:0021954 |
central nervous system neuron development(GO:0021954) |
| 0.0 |
0.3 |
GO:0001845 |
phagolysosome assembly(GO:0001845) |
| 0.0 |
0.6 |
GO:0071774 |
response to fibroblast growth factor(GO:0071774) |
| 0.0 |
0.1 |
GO:0032367 |
intracellular cholesterol transport(GO:0032367) |
| 0.0 |
0.3 |
GO:0016926 |
protein desumoylation(GO:0016926) |
| 0.0 |
0.0 |
GO:0035793 |
negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) cell migration involved in kidney development(GO:0035787) cell migration involved in metanephros development(GO:0035788) metanephric mesenchymal cell migration(GO:0035789) positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) regulation of metanephric mesenchymal cell migration(GO:2000589) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.0 |
0.2 |
GO:0061099 |
negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 |
0.1 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 |
0.1 |
GO:0070537 |
histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 |
0.3 |
GO:0016572 |
histone phosphorylation(GO:0016572) |
| 0.0 |
0.6 |
GO:0034314 |
Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 |
0.1 |
GO:0070932 |
histone H3 deacetylation(GO:0070932) |
| 0.0 |
0.1 |
GO:0050861 |
positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 |
0.8 |
GO:0045667 |
regulation of osteoblast differentiation(GO:0045667) |
| 0.0 |
0.1 |
GO:0035414 |
negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 |
0.5 |
GO:0051453 |
regulation of intracellular pH(GO:0051453) |
| 0.0 |
0.1 |
GO:0016056 |
rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 |
0.2 |
GO:0051988 |
regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 |
0.4 |
GO:0045760 |
positive regulation of action potential(GO:0045760) |
| 0.0 |
0.2 |
GO:1903608 |
protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 |
1.0 |
GO:0060999 |
positive regulation of dendritic spine development(GO:0060999) |
| 0.0 |
0.2 |
GO:0019368 |
fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 |
0.1 |
GO:0000289 |
nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 |
0.1 |
GO:0035507 |
regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 |
0.1 |
GO:0060236 |
regulation of mitotic spindle organization(GO:0060236) regulation of spindle organization(GO:0090224) |
| 0.0 |
0.2 |
GO:0000712 |
resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 |
0.2 |
GO:0044342 |
type B pancreatic cell proliferation(GO:0044342) |
| 0.0 |
0.2 |
GO:1901621 |
negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 |
0.6 |
GO:0006754 |
ATP biosynthetic process(GO:0006754) |
| 0.0 |
0.9 |
GO:0046513 |
ceramide biosynthetic process(GO:0046513) |
| 0.0 |
0.1 |
GO:0002949 |
tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 |
0.2 |
GO:1900006 |
positive regulation of dendrite development(GO:1900006) |
| 0.0 |
0.1 |
GO:1903587 |
regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.0 |
0.1 |
GO:0001672 |
regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 |
0.1 |
GO:0046501 |
protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 |
0.4 |
GO:0002089 |
lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 |
0.1 |
GO:0009301 |
snRNA transcription(GO:0009301) |
| 0.0 |
0.4 |
GO:0080171 |
lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 |
0.1 |
GO:0030263 |
apoptotic chromosome condensation(GO:0030263) |
| 0.0 |
0.1 |
GO:0031954 |
positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 |
0.1 |
GO:0006903 |
vesicle targeting(GO:0006903) |
| 0.0 |
0.1 |
GO:0033275 |
actin-myosin filament sliding(GO:0033275) |
| 0.0 |
0.5 |
GO:0035640 |
exploration behavior(GO:0035640) |
| 0.0 |
0.1 |
GO:0060527 |
prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.0 |
0.1 |
GO:0070124 |
mitochondrial translational initiation(GO:0070124) |
| 0.0 |
0.7 |
GO:0090090 |
negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 |
0.1 |
GO:0043461 |
proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 |
0.1 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.0 |
0.2 |
GO:0044804 |
nucleophagy(GO:0044804) |
| 0.0 |
0.1 |
GO:0006167 |
AMP biosynthetic process(GO:0006167) |
| 0.0 |
0.1 |
GO:0071947 |
protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 |
0.1 |
GO:0035024 |
negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 |
0.1 |
GO:0010667 |
negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 |
0.2 |
GO:0015732 |
prostaglandin transport(GO:0015732) |
| 0.0 |
0.2 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 |
0.1 |
GO:1902042 |
negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 |
0.0 |
GO:0045002 |
double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 |
0.1 |
GO:0070193 |
synaptonemal complex organization(GO:0070193) |
| 0.0 |
0.7 |
GO:0035418 |
protein localization to synapse(GO:0035418) |
| 0.0 |
0.1 |
GO:0006390 |
transcription from mitochondrial promoter(GO:0006390) |
| 0.0 |
0.0 |
GO:0070126 |
mitochondrial translational termination(GO:0070126) |
| 0.0 |
0.0 |
GO:0006558 |
L-phenylalanine metabolic process(GO:0006558) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) |
| 0.0 |
0.1 |
GO:0007021 |
tubulin complex assembly(GO:0007021) |
| 0.0 |
0.1 |
GO:0097368 |
establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 |
0.1 |
GO:0051189 |
Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 |
0.0 |
GO:1902605 |
heterotrimeric G-protein complex assembly(GO:1902605) |
| 0.0 |
0.1 |
GO:0000480 |
endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 |
0.3 |
GO:0051310 |
metaphase plate congression(GO:0051310) |
| 0.0 |
0.1 |
GO:0050832 |
defense response to fungus(GO:0050832) |
| 0.0 |
0.0 |
GO:0035526 |
retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 |
0.1 |
GO:0060394 |
negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 |
0.3 |
GO:0006474 |
N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 |
0.2 |
GO:0070536 |
protein K63-linked deubiquitination(GO:0070536) |
| 0.0 |
0.1 |
GO:0045086 |
positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 |
0.1 |
GO:0046950 |
cellular ketone body metabolic process(GO:0046950) |
| 0.0 |
0.1 |
GO:0010457 |
centriole-centriole cohesion(GO:0010457) |
| 0.0 |
0.0 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 |
0.0 |
GO:0019043 |
establishment of viral latency(GO:0019043) |
| 0.0 |
0.2 |
GO:0045722 |
positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 |
0.1 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) |
| 0.0 |
0.0 |
GO:0060011 |
Sertoli cell proliferation(GO:0060011) |
| 0.0 |
0.0 |
GO:0070278 |
extracellular matrix constituent secretion(GO:0070278) |
| 0.0 |
0.1 |
GO:1904294 |
positive regulation of ERAD pathway(GO:1904294) |
| 0.0 |
0.3 |
GO:0046470 |
phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 |
0.1 |
GO:2000114 |
regulation of establishment of cell polarity(GO:2000114) |
| 0.0 |
0.3 |
GO:0038083 |
peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 |
0.0 |
GO:0007084 |
mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 |
0.1 |
GO:0006968 |
cellular defense response(GO:0006968) |
| 0.0 |
0.1 |
GO:0006359 |
regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 |
0.1 |
GO:0007263 |
nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 |
0.1 |
GO:0007340 |
acrosome reaction(GO:0007340) |
| 0.0 |
0.1 |
GO:0032808 |
lacrimal gland development(GO:0032808) |
| 0.0 |
0.1 |
GO:0051085 |
chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 |
0.4 |
GO:0030433 |
ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 |
0.4 |
GO:0032755 |
positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 |
0.2 |
GO:0070584 |
mitochondrion morphogenesis(GO:0070584) |
| 0.0 |
0.5 |
GO:0042147 |
retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 |
0.1 |
GO:0070884 |
regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 |
0.7 |
GO:0007601 |
visual perception(GO:0007601) |
| 0.0 |
0.6 |
GO:0032543 |
mitochondrial translation(GO:0032543) |
| 0.0 |
0.1 |
GO:0001914 |
regulation of T cell mediated cytotoxicity(GO:0001914) |
| 0.0 |
0.2 |
GO:0001755 |
neural crest cell migration(GO:0001755) |
| 0.0 |
0.1 |
GO:0010575 |
positive regulation of vascular endothelial growth factor production(GO:0010575) |
| 0.0 |
0.2 |
GO:0034340 |
response to type I interferon(GO:0034340) |
| 0.0 |
0.5 |
GO:0006506 |
GPI anchor biosynthetic process(GO:0006506) |
| 0.0 |
0.0 |
GO:0070493 |
thrombin receptor signaling pathway(GO:0070493) |
| 0.0 |
0.1 |
GO:0021521 |
ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 |
0.3 |
GO:0001658 |
branching involved in ureteric bud morphogenesis(GO:0001658) |
| 0.0 |
0.3 |
GO:0070527 |
platelet aggregation(GO:0070527) |
| 0.0 |
0.0 |
GO:0001779 |
natural killer cell differentiation(GO:0001779) |