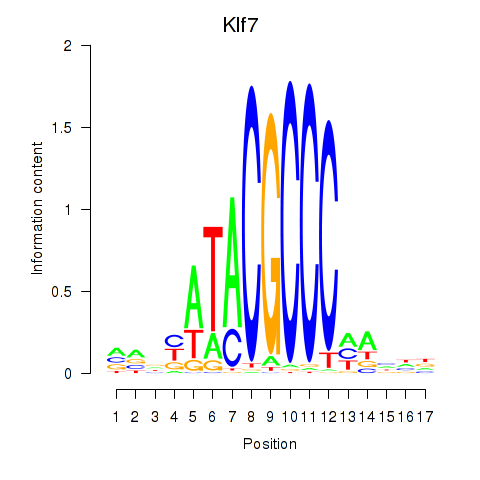
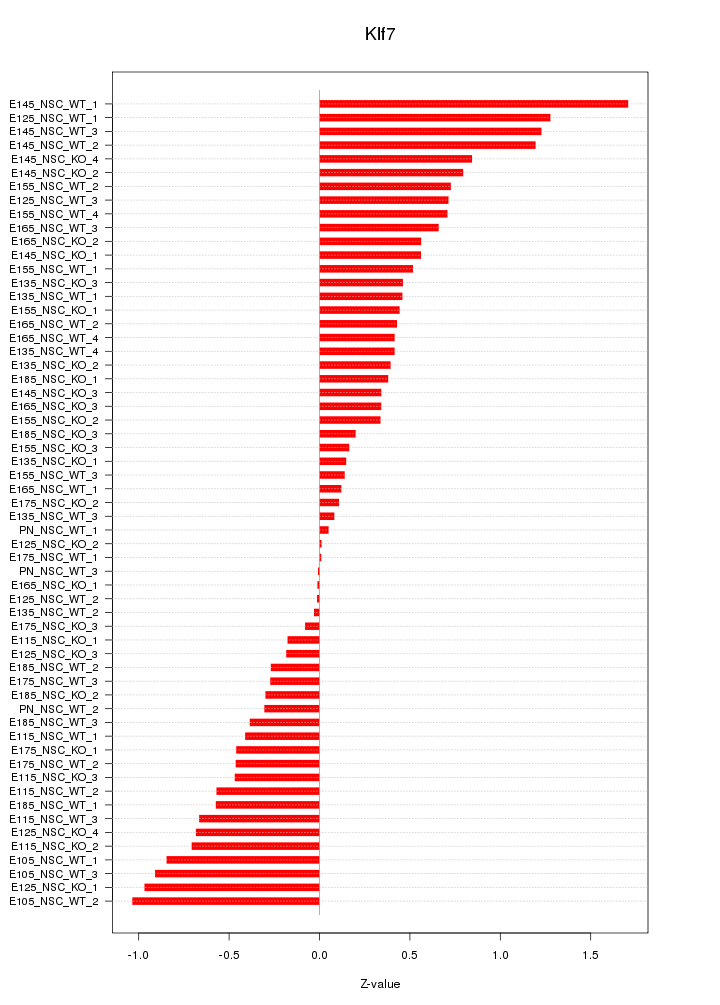
| 1.8 |
5.5 |
GO:0021852 |
pyramidal neuron migration(GO:0021852) |
| 1.0 |
3.1 |
GO:0032915 |
positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.8 |
4.2 |
GO:0002457 |
T cell antigen processing and presentation(GO:0002457) |
| 0.8 |
3.8 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.5 |
8.0 |
GO:0050901 |
leukocyte tethering or rolling(GO:0050901) |
| 0.5 |
1.4 |
GO:0070375 |
ERK5 cascade(GO:0070375) |
| 0.3 |
2.0 |
GO:0007253 |
cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.3 |
3.0 |
GO:0038026 |
reelin-mediated signaling pathway(GO:0038026) |
| 0.3 |
1.2 |
GO:0061428 |
positive regulation of oocyte development(GO:0060282) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.2 |
0.9 |
GO:1900533 |
medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.2 |
1.5 |
GO:0043983 |
histone H4-K12 acetylation(GO:0043983) |
| 0.2 |
1.3 |
GO:0006537 |
glutamate biosynthetic process(GO:0006537) proline biosynthetic process(GO:0006561) |
| 0.2 |
0.6 |
GO:1900110 |
negative regulation of histone H3-K9 dimethylation(GO:1900110) |
| 0.2 |
3.0 |
GO:0001574 |
ganglioside biosynthetic process(GO:0001574) |
| 0.1 |
1.5 |
GO:0007191 |
adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 |
0.9 |
GO:0035845 |
photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 |
0.7 |
GO:1901837 |
negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.1 |
0.5 |
GO:1903599 |
attachment of mitotic spindle microtubules to kinetochore(GO:0051315) positive regulation of mitophagy(GO:1903599) |
| 0.1 |
0.3 |
GO:0046341 |
CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 |
0.5 |
GO:0035262 |
gonad morphogenesis(GO:0035262) |
| 0.1 |
0.5 |
GO:0006451 |
selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 |
0.3 |
GO:0051151 |
negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 |
0.2 |
GO:0035621 |
ER to Golgi ceramide transport(GO:0035621) ceramide transport(GO:0035627) |
| 0.0 |
0.7 |
GO:0060628 |
regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 |
0.1 |
GO:0060598 |
dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.0 |
0.2 |
GO:0015803 |
branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 |
3.3 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.0 |
1.2 |
GO:0051443 |
positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 |
3.5 |
GO:0014065 |
phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.0 |
0.5 |
GO:0030157 |
pancreatic juice secretion(GO:0030157) |
| 0.0 |
0.5 |
GO:0019243 |
methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 |
0.9 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
4.1 |
GO:0030518 |
intracellular steroid hormone receptor signaling pathway(GO:0030518) |
| 0.0 |
2.1 |
GO:0032088 |
negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 |
0.3 |
GO:0097034 |
mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 |
1.9 |
GO:0060997 |
dendritic spine morphogenesis(GO:0060997) |
| 0.0 |
1.4 |
GO:0006970 |
response to osmotic stress(GO:0006970) |
| 0.0 |
0.5 |
GO:0097352 |
autophagosome maturation(GO:0097352) |
| 0.0 |
1.2 |
GO:0032543 |
mitochondrial translation(GO:0032543) |
| 0.0 |
0.8 |
GO:0060292 |
long term synaptic depression(GO:0060292) |
| 0.0 |
0.7 |
GO:0035774 |
positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 |
0.0 |
GO:0031590 |
wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.0 |
0.1 |
GO:0071318 |
cellular response to ATP(GO:0071318) |
| 0.0 |
0.2 |
GO:0016558 |
protein import into peroxisome matrix(GO:0016558) |